How iota2 is designed
Introduction
Originally, iota2 was designed as a classification workflow for large scale land cover mapping. As such, it includes a lot of tools from data preprocessing up to the metric computation from a confusion matrix. Among all these tools, many of them are not specific to supervised classification and can be useful for othe data generation purposes (regression, feature extraction, etc.) as long as they need to be applied to several tiles in a scalable way.
How is iota2 built
iota2 is built using three concepts:
These three elements allow to manage large amounts of data, and several combinations of inputs.
Tasks
A task corresponds to the processing applied to some inputs which produces output data which can be fed to the next task or can provide a final product like a land cover map.
For instance, in a classification workflow, a task can correspond to computing the cloud cover for a given tile, computing the ratio of samples, training a model or merging several classifications.
A task may be run many times for a range of parameters, e.g. a tile, a set of dates, a climatic region, etc.
Steps
A step is a container which manages several tasks which can be run together in sequence. The main goal of a step is to perforn all linked tasks. A set of tasks is put together into a step for computational purposes. For instance some tasks, when connected together, can pass data to one another in memory without needing to write intermediate outputs to disk. Using steps allows to divide the whole pipeline in several independent parts. This way, if errors occur during the execution, the completed steps don’t need to be run again. Once the error is corrected, the execution can be resumed starting with the first uncomplete step.
The step manages all the parameters and distributes them to the different tasks. It is also in charge of defining how many times each task is called.
More information about steps are available here
Groups
A group contains several steps. Unlike steps, a group has a more abstract value because it is only used for scheduling purposes. A group is used to label a set of steps. The iota2 parameters then allow to access the different groups to perform only a part of the workflow.
Simplified classification workflow
The following example illustrates the link between tasks, steps and groups.
To this end, a very simplified workflow is considered:
Compute the common mask between all images for each tile
Compute a binary mask for clouds, saturations and borders
Compute the class ratio between learning and validation
Extract the samples for learning
Train the classifier
Classify the images
Merge all the classifications to produce an unique map
![digraph {
rankdir=TB;
subgraph cluster_0 {
color=green;
label="Group: Init";
a [label= "Common Mask", shape=box, color=blue]
t11 [label="tile 1"]
t12 [label="tile 2"]
a -> t11 [style=dotted];
a-> t12 [style=dotted];
b [label="Cloud Cover", shape=box, color=blue]
a -> b;
t1n1 [label="tile N-1"]
t1n [label="tile N"]
a -> t1n1 [style=dotted];
a -> t1n [style=dotted];
t112 [label="tile 1"]
t122 [label="tile 2"]
t1n12 [label="tile N-1"]
t1n2 [label="tile N"]
b -> t112 [style=dotted];
b -> t122 [style=dotted];
b -> t1n12 [style=dotted];
b -> t1n2 [style=dotted];
}
Tasks
Steps [color=blue, shape=box]
Groups [color=green, shape=box]
subgraph cluster_1 {
color=green;
label="Group: Sampling";
c [label="Samples selection", shape=box, color=blue]
b -> c;
t21 [label="tile 1"]
t22 [label="tile 2"]
c -> t21 [style=dotted];
c-> t22 [style=dotted];
d [label="Samples Extraction", shape=box, color=blue]
c -> d;
t2n1 [label="tile N-1"]
t2n [label="tile N"]
c -> t2n1 [style=dotted];
c -> t2n [style=dotted];
t212 [label="tile 1"]
t222 [label="tile 2"]
t2n12 [label="tile N-1"]
t2n2 [label="tile N"]
d -> t212 [style=dotted];
d -> t222 [style=dotted];
d -> t2n12 [style=dotted];
d -> t2n2 [style=dotted];
}
subgraph cluster_2 {
color=green;
label="Learning and Classification";
e [label="Learning", shape=box, color=blue]
d -> e;
t31 [label="Region 1"]
e -> t31 [style=dotted];
f [label="Classification", shape=box, color=blue]
e -> f;
t312 [label="tile 1"]
t322 [label="tile 2"]
t3n12 [label="tile N-1"]
t3n2 [label="tile N"]
f -> t312 [style=dotted];
f -> t322 [style=dotted];
g [label="Merge tiles", color=blue, shape=box]
f -> t3n12 [style=dotted];
f -> t3n2 [style=dotted];
f->g;
g -> "All tiles";
}
}](_images/graphviz-852b17891540b2f654a68f4df1dc072e5fe11c1c.png)
What is a builder ?
The sequence of steps as illustrated above represents a builder. It represents the order in which the different steps are performed.
The aim of generic builders, introduced here is to encourage users to create their own chains, using the iota2 API.
What is contained in a builder ?
A builder builds the task graph, in other terms, it creates a chain of several Steps.
To allow custom builders, inheritance is used.
The superclass called i2_builder, contains all generic methods used by iota2 for logging or printing workflows.
It defines also the most important function: build_steps which must be implemented in each derived builder.
The __init__ function
Each builder is composed of two mandatory attributes:
steps_groups: an ordered dict. Each key is an unique string corresponding to a group name. Each key stores an ordered dict dedicated to a group. Each group contains an ordered set of steps.steps: an ordered list of all steps to be done. This list is filled using the class methodbuild_steps.
The build_steps function:
This is the central point of a user contribution to create a new chain.
This function is simply a concatenation of all steps the order in which they need to be processed.
Each step is initialized, and added to a StepContainer which is returned at the end of build_steps function.
It is strongly recommended to add checks on the configuration file variables and also to verify if steps aro allowed to be launch in the same run. This is very important especially if several combination of steps are allowed. For a concrete example, refer to I2Classification builder.
Use a new chain
With this information, it is possible to create a new chain by simply creating a new builder.
To indicate which builder must be used when iota2 is launched, it is necessary to fill the configuration file in the dedicated block: builders
Field name |
Type |
Required |
builders_paths |
string |
Optional |
builders_class_name |
list |
Optional |
All these parameters are optional, as by default the builder used produces the land cover map over the specified tiles.
builders_paths: locate where builders are stored. If not indicated, the chain will look into the relative path IOTA2DIR/iota2/sequence_builderbuilders_class_name: the builders class name to use, ordered in a list. If not indicated, the chain will launch the classification builder. A .py file can contain several classes or builders. Choosing the one to be used is mandatory. Available builders are : I2Classification and I2FeaturesMap.
If one of these parameters is not consistent with the others, the chain will return an error.
Example
In this section, a new chain will be designed and created entirely. The goal here is to provide a working example, using both new and existing functions from the iota2 API.
Design the new chain
This chain simulates the classification of images based on segmentation.
To do this, the chain must perform the initial segmentation, then calculate zonal statistics that will be used to train a classifier. Then, still by exploiting the zonal statistics and the learned model, a classification is carried out, providing the final product expected by the chain.
To simplify the construction of the chain and provide a very fast execution, the data are replaced by text files, and the different algorithms will modify the content of these files. However, the programming paradigms of iota2 will be respected, such as tile processing for example.
Creating the chain
The first thing to do is to code the functions that will be used to perform the processing. Most of the functions presented in this section perform unnecessary or pointless operations. The most important being the construction of the steps and the dependency graph. To write these functions we have to keep in mind the granularity of the processing we want to apply.
Functions definition
The first function usually consists in creating the tree structure of the output folders. This one requires only one input parameter: a path to a directory.
def create_directories(root: str) -> None:
"""
Function for creating folders
The tree structure never changes, because the folder names are used
in the different steps.
Parameters
----------
root:
the high level output path
"""
if Path(root).exists():
shutil.rmtree(root)
Path(root).mkdir()
folders = ["segmentation", "statistics", "model", "classif", "merge_tiles", "final"]
for folder in folders:
Path(root, folder).mkdir(exist_ok=True, parents=True)
The second step is to perform the segmentation. In this example, we simulate a tile segmentation by writing a random number of segments. This trick is realistic because when segmenting a satellite image, the number of detected objects cannot be known in advance. Whereas the previous function only needed a directory, for this one we perform the processing only for one tile. When using Sentinel-2 data over a whole year, it is not possible to hold several tiles in memory at the same time.
def write_segments_by_tiles(output_path: str, tile: str, seed: int = 0) -> None:
"""
Write a file containing a list of segments.
The number of segment is random.
Parameters
----------
tile :
A tile name
seed :
Fix the random seed
"""
folder = str(Path(output_path) / "segmentation")
# fix the random seed for reproducibility,
if seed != 0:
random.seed(seed)
# pick a random number between 0 and 50
rand = random.randint(1, 11)
with open(str(Path(folder) / f"{tile}.txt"), "w", encoding="utf-8") as tile_file:
chain = [f"Segment_{tile}_{i}" for i in range(int(rand))]
tile_file.write("\n".join(chain))
Once the segmentation is completed, we can compute zonal statistics that will be used for training and classification. This processing depends again on a tile, and provides a result for each segment present in the tile.
def compute_zonal_stats(output_path: str, tile: str) -> None:
"""
For a given tile, simulate the zonal stats computation.
"""
in_folder = str(Path(output_path) / "segmentation")
out_folder = str(Path(output_path) / "statistics")
with open(
str(Path(out_folder) / f"Stats_{tile}.txt"), "w", encoding="utf-8"
) as new_file:
with open(str(Path(in_folder) / f"{tile}.txt"), encoding="utf-8") as file_tile:
contents = file_tile.read().split("\n")
for line in contents:
chain = line.replace("Segment", "Stats") + "\n"
new_file.write(chain)
Now, we can move on to training the classifier. To do this, we will search all the files containing the statistics. The training here is fake, we simply count the number of files found.
def training_model(output_path: str) -> None:
"""
Simulate the training of a classifier
"""
model_file = str(Path(output_path) / "model" / "Model_global.txt")
# get all stats files
stats_files = FSA(str(Path(output_path) / "statistics"), True, "Stats", ".txt")
with open(model_file, "w", encoding="utf-8") as model:
model.write(str(len(stats_files)))
Once the model has been trained, we can move on to classification. For this example, a constraint of the classifier is that it can only process one segment at a time. So we need to call this classifier for each segment.
def classify(output_path: str, tile: str, segment_number: int) -> None:
"""
For a given tile, simulate the classification of each segment.
This classifier algorithm is able to classify only one segment at the
same time and need to write the output in a temporary file.
"""
in_folder = str(Path(output_path) / "statistics")
out_folder = str(Path(output_path) / "classif")
model = str(Path(output_path) / "model" / "Model_global.txt")
with open(
str(Path(in_folder) / f"Stats_{tile}.txt"), encoding="utf-8"
) as file_tile:
contents = file_tile.read().split("\n")
for cpt, line in enumerate(contents):
if cpt == segment_number:
with open(
str(Path(out_folder) / f"classif_{tile}_{cpt}.txt"),
"w",
encoding="utf-8",
) as sub_file:
chain = f"{model} " + line.replace("Stats", "Classif") + "\n"
sub_file.write(chain)
Once the classification is finished, we will gather the different segments, first by tile and then in a single file all tiles together.
def merge_classification_by_tile(output_path: str, tile: str) -> None:
"""
Method used in merge_classification_by_tile example step. Write all content of classif text
files in a single text file (for a given tile).
Parameters
----------
output_path:
Path to the output directory
tile:
Tile name
"""
# get all files created for this tile
in_folder = str(Path(output_path) / "classif")
out_folder = str(Path(output_path) / "merge_tiles")
files = FSA(in_folder, True, f"classif_{tile}_", ".txt")
files.sort()
with open(
str(Path(out_folder) / f"result_{tile}.txt"), "w", encoding="utf-8"
) as outfile:
for fil in files:
with open(fil, encoding="utf-8") as infile:
outfile.write(infile.read())
def mosaic(output_path: str) -> None:
"""
Method used in mosaic example step. Write all content of .txt files in the final directory in a
single file.
Parameters
----------
output_path:
Path to the output directory
"""
in_folder = str(Path(output_path) / "merge_tiles")
out_folder = str(Path(output_path) / "final")
files = FSA(in_folder, True, ".txt")
files.sort()
with open(str(Path(out_folder) / "final.txt"), "w", encoding="utf-8") as outfile:
for fil in files:
with open(fil, encoding="utf-8") as infile:
outfile.write(infile.read())
Now we have all the functions required to produce a classification from a segmentation. We need to link them to produce the chain. This is done in two phases, the creation of Steps and the creation of builder.
Step declaration
To start writing the steps, you have to keep in mind their sequence, and especially the dependencies between steps.
The general workflow is given by the previous section since the functions are listed in logical sequence. In this section the emphasis will be on the links between the steps.
The first step is to create the tree structure. So we will write a step that calls the corresponding function.
Several important points:
resources_block_name is a mandatory attribute, it allows you to allocate resources to a particular processing. Indeed, creating directories is less consuming than computing a segmentation.
the call to i2_task allows to provide parameters to the function called in the step. It is important to make sure that all the mandatory parameters are filled in at this stage.
add_task_to_i2_processing_graph is the function that adds the different steps to the Dask scheduler.
finally, the class method step_description allows a better readability of the different graphical representations of the chain that we will see later.
As this is the first step, there are no dependencies to be expressed. The second step is writing the segmentation. We can note that we define as many tasks as tiles, and each task is added to the graph. An important point here concerns the task_name, it must depend on the tile name because each task_name must be unique in the execution graph.
This step can start once the previous one is done. It’s expressed through task_dep_dico={“first_task”:[]}. This means that we wait until all items in the list corresponding to first_task are completed. Here we indicate that each task has its own label, with task_sub_group, which depends on the tile name.
Once the segmentation is done, the zonal statistics can be computed for each tile which has already been processed. Then the step looks like the following.
Again, the dependencies are expressed as a dictionary containing a list. In this case one simply waits until the corresponding tile has been processed.
To train the model, we wait for all the tiles to be processed. The corresponding step expresses the dependencies by waiting for all zonal statistics tasks.
Until now, the dependencies between the different steps were set at the level of the tile. For the next step, we need to know the number of segments per tile. Due to the constraint we impose on the classifier, the number of segmentations per tile cannot be known before the calculation of the segmentation . To overcome this problem we need to divide the execution graph in two. This separation could have occurred at any time between the calculation of the segmentation and the classification. We have made this choice in order to illustrate several cases of dependencies. The following figure represents the execution graph, from the creation of the directories to the learning of the model.
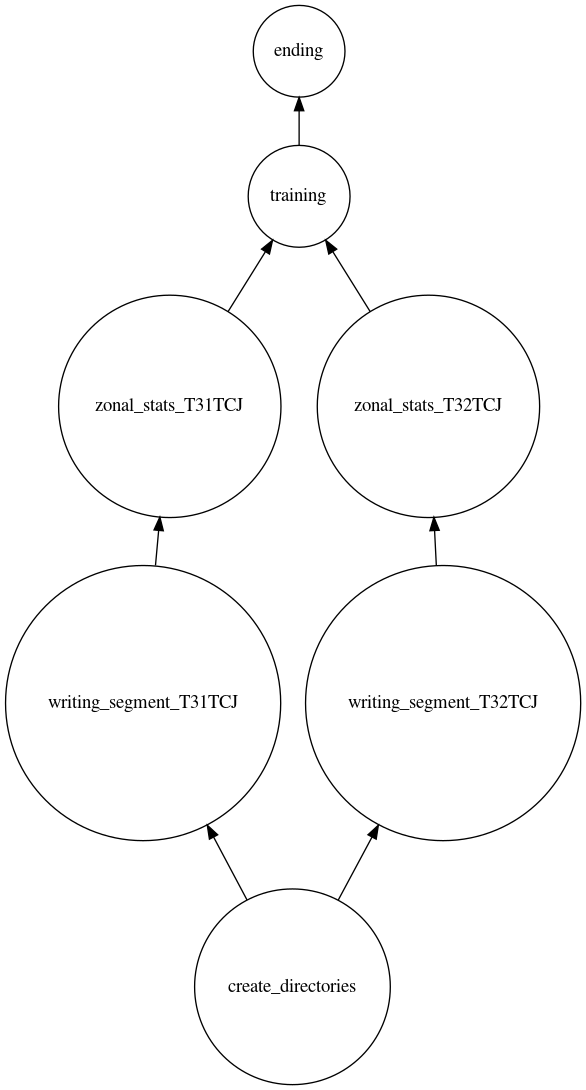
To perform the classification and merge steps, we need to create a second execution graph.
This way the classification step does not depend on any previous step. We will see later how to express the dependency between the two graphs.
The classification step has an empty dependency list, but generates dependencies that are named using both the tile name and the segment identifier.
Once the classification of a tile is finished we can carry out the merging of the different segments.
Finally, when all the segments of all the tiles have been merged by tile, we can make the mosaic of the different tiles.
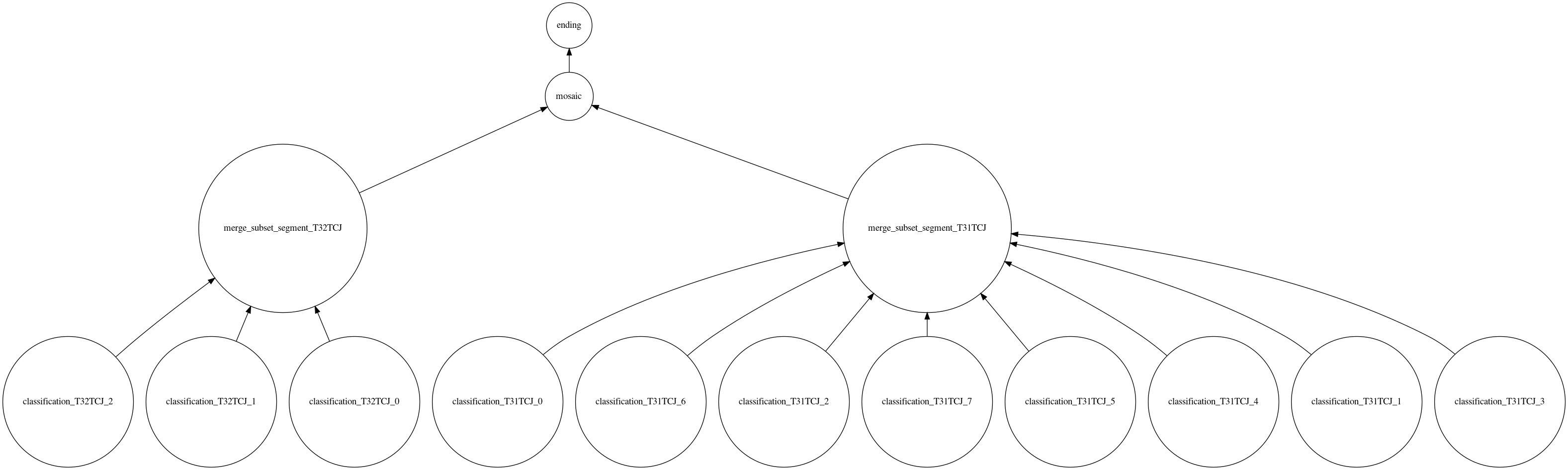
At this stage, we have created all the necessary steps for the development of the processing chain. We have expressed the dependencies between the different steps. However, we still lack the central part to make iota2 understand that this is a new processing chain. It is thus necessary to write the builder.
Writing the builder
A builder is a class which inherits from i2_builder.
First the __init__ method, instantiates attributes and defines groups. Groups are only labels allowing to restart the chain for a specific block. For this example, we have only one group named global.
The function build_steps is the most important. As we need two graphs, we define two StepContainer. We add steps to the container. As we need to know how many segments we have by tile, we use the function get_number_of_segment_by_tiles, which fills a dictionary containing the number of segments for each tile. This dictionary is filled before the chain instantiates the second container, after the execution of all steps of the first StepContainer.
Then the complete builder looks like this:
Congratulations, the new chain is created, but before using it, we must fill the corresponding configuration file.
Filling a configuration file
In iota2, all the parameters defined and used are known by all builders. When creating a new builder, it can be useful to use the existing parameters instead of creating new ones. For this example, we use existing blocks like chain or builder which are mandatory, and create a new one MyParams. In MyParams we define a parameter tiles which is identical to existing listTile in order to show that there is no conflict between different blocks.
chain:
{
output_path:"/absolute_path/to/ouput_dir"
first_step:"global"
last_step:"global"
}
MyParams:
{
tiles:"T31TCJ T32TCJ"
seed:0
}
builders:
{
builder_file_path:"/home/myBuilders_python_packages"
builder_class_name:["i2_example_builder"]
}
Warning
Iota2 is able to merge builders into one by the use of workflow_merger. Currently, builders sharing the same step cannont be merged.