Object Based Image Analysis (OBIA)
To perform OBIA, the first need is to define “objects”. The objects can be automatically detected using segmentation algorithms, or can be defined by the user as for instance using agricultural plots from field campaigns or databases such as the Land Parcel Information System.
For the OBIA approach, it is necessary to compute features for each object. In this implementation, five zonal statistics can be computed, and used together, for each band or spectral index: mean, minimum, maximum, standard deviation or count (number of pixels in the object). For this, all pixels in the objects are used.
Once features are computed, classification algorithms can be used. Training and classification steps are done at object scale.
Using objects instead of pixels raises particular difficulties, especially on multi-tile configurations. See the technical documentation for more information about this.
Introduction to data
iota2 handles several sensors:
Landsat 5 and 8 (old and new THEIA format)
Sentinel-1, Sentinel-2 L2A (THEIA and Sen2cor), Sentinel-2 L3A (Theia Format)
Various other images already processed, with the
userFeatsensor (which is not a real sensor)
In this chapter, only the use of Sentinel-2 L2A will be illustrated. To use other sensors, it is necessary to adapt the input parameters according to parameters descriptions.
iota2 uses machine learning algorithms to produce land cover maps. It requires, among others inputs, images and related reference data.
Get the data set
Two data-set are available, containing minimal data required to run Iota2 builders:
A entire Sentinel 2 tile, with two dates (8.8 Go)
An extraction of Sentinel 2 data, with three dates over different eco-climatic region (Soon)
The archive contains:
- /XXXX/IOTA2_TEST_S2
- archive content
content of the tutorial archive after content extraction
- ! external_code
- python code folder
user custom python code used for external features / feature maps
- external_code.py
- python code file
contains user python code used to produce the spectral indices. Rules for creating user code are explained in external features page.
- ! IOTA2_Outputs
- output folder
folder used for iota2 output folders
(empty)
- sensor_data
- input raster data
the directory which contains Sentinel-2 data. These data must be stored by tiles as in the archive.
- T31TCJ
- ! SENTINEL2A_20180511-105804-037_L2A_T31TCJ_D_V1-7
- MASKS
SENTINEL2A_20180511-105804-037_L2A_T31TCJ_D_V1-7_*.tif
SENTINEL2A_20180511-105804-037_L2A_T31TCJ_D_V1-7_FRE_B*.tif
SENTINEL2A_20180511-105804-037_L2A_T31TCJ_D_V1-7_FRE_STACK.tif
- ! SENTINEL2A_20180521-105702-711_L2A_T31TCJ_D_V1-7
- MASKS
SENTINEL2A_20180521-105702-711_L2A_T31TCJ_D_V1-7_*.tif
SENTINEL2A_20180521-105702-711_L2A_T31TCJ_D_V1-7_FRE_B*.tif
SENTINEL2A_20180521-105702-711_L2A_T31TCJ_D_V1-7_FRE_STACK.tif
- T31TDJ
- ! SENTINEL2A_20180511-105804-037_L2A_T31TDJ_D_V1-7
- MASKS
SENTINEL2A_20180511-105804-037_L2A_T31TDJ_D_V1-7_*.tif
SENTINEL2A_20180511-105804-037_L2A_T31TDJ_D_V1-7_ATB_R1.tif
SENTINEL2A_20180511-105804-037_L2A_T31TDJ_D_V1-7_FRE_B*.tif
SENTINEL2A_20180511-105804-037_L2A_T31TDJ_D_V1-7_FRE_STACK.tif
SENTINEL2A_20180511-105804-037_L2A_T31TDJ_D_V1-7_MTD_ALL.xml
SENTINEL2A_20180511-105804-037_L2A_T31TDJ_D_V1-7_QKL_ALL.jpg
- ! SENTINEL2A_20180521-105702-711_L2A_T31TDJ_D_V1-7
- MASKS
SENTINEL2A_20180521-105702-711_L2A_T31TDJ_D_V1-7_*.tif
SENTINEL2A_20180521-105702-711_L2A_T31TDJ_D_V1-7_ATB_R1.tif
SENTINEL2A_20180521-105702-711_L2A_T31TDJ_D_V1-7_FRE_B*.tif
SENTINEL2A_20180521-105702-711_L2A_T31TDJ_D_V1-7_FRE_STACK.tif
SENTINEL2A_20180521-105702-711_L2A_T31TDJ_D_V1-7_MTD_ALL.xml
SENTINEL2A_20180521-105702-711_L2A_T31TDJ_D_V1-7_QKL_ALL.jpg
- vector_data
- input vector data
directory containing input vector data
reference_data.cpg
reference_data.dbf
reference_data.prj
- reference_data.shp
- shapefile
the shapeFile containing geo-referenced and labelled polygons (no multi-polygons, no overlapping) used to train a classifier.
reference_data.shx
EcoRegion.dbf
EcoRegion.prj
EcoRegion.qpj
- EcoRegion.shp
- shapefile
shapeFile containing two geo-referenced polygons representing a spatial stratification (eco-climatic areas, for instance).
EcoRegion.shx
- colorFile.txt
- color table
colors used in classification map
$ cat colorFile.txt ... 211 255 85 0 ...
Here the class 211 has the RGB color 255 85 0.
- IOTA2_Example.cfg
- example config file
the file used to set iota2’s parameters such as inputs/outputs paths, classifier parameters etc.
- i2_tutorial_classification.cfg
- sample config file
sample config file for classification builder
- i2_tutorial_features_map.cfg
- sample config file
sample config file for feature map builder
- i2_tutorial_obia.cfg
- sample config file
sample config file for object base image analysis
- nomenclature23.txt
- nomenclature file
label’s name. The purpose of the file is to get a pretty results report at the end of the chain by relabeling integers labels by a more verbose type.
& cat nomenclature.txt ... prairie:211 ...
Here the class 211 corresponds to the class prairie
vecteur_23.qml
Understanding the configuration file
iota2 uses hundreds of parameters, some of them are specfic to iota2 and others come from other libraries such as scikit-learn or OTB.
These parameters allow selecting the processing steps to be carried out and their parameters. A documentation of all these parameters is provided. The user defines these paramereters in a configuration file (a human readable text file) that is read by iota2 at start-up. The file is structured into sections, each section containing various fields.
A section (or group of fields) contains fields with similar purposes. For instance, the section chain contains general information (such as input data, the output path,…) and the section arg_train will contain information about the classifier parameters (name of classifier, number of tree for Random Forest or cost for Support Vector Margin).
This minimal configuration file contains all required fields for OBIA:
chain :
{
output_path : 'XXXX/IOTA2_TEST_S2/IOTA2_Outputs/Results-obia'
remove_output_path : True
nomenclature_path: "XXXX/IOTA2_TEST_S2/nomenclature23.txt"
list_tile : 'T31TCJ'
s2_path : 'XXXX/IOTA2_TEST_S2/sensor_data'
ground_truth: "XXXX/IOTA2_TEST_S2/vector_data/groundTruth.shp"
data_field : 'code'
spatial_resolution : 10
color_table : "XXXX/IOTA2_TEST_S2/colorFile.txt"
proj : 'EPSG:2154'
first_step: "init"
last_step: "validation"
}
arg_train :
{
classifier : 'sharkrf'
otb_classifier_options:{
"classifier.sharkrf.nbtrees" : 100
}
}
arg_classification :
{
classif_mode : 'separate'
}
obia:
{
buffer_size: 2000
}
builders:
{
builders_class_name: ["I2Obia"]
}
Among these parameters, some require a particular attention:
output_path : path to write results
data_field : the label column name in the reference data
buffer_size : this parameter manages the RAM consumption of obiasteps. The RAM used is proportional to the number of dates, the number of statistics used, and the images size.
sample_selection : this parameters allows to manage the class proportions. In OBIA, the proportions are computed by the number of polygons, not area or number of pixels.
spatial_resolution : the spatial resolution must be consistent with the size of object in segmentation. Statistics can be computed only if objects are larger than pixels.
For an end user, running iota2 requires correctly filling the configuration file.
In the above example, replace the XXXX by the path where the archive has been extracted.
Running the chain
iota2 launch
The chain is launched with the following command line.
Iota2.py -config /XXXX/IOTA2_TESTS_DATA/i2_tutorial_obia.cfg -scheduler_type localCluster
First, the chain displays the list of all steps activated by the configuration file.
Group init:
[x] Step 1: check inputs
[x] Step 2: Sensors pre-processing
[x] Step 3: Generate a common masks for each sensors
[x] Step 4: Compute validity raster by tile
[x] Step 5: Compute SLIC segmentation by tile
[x] Step 6: Vectorize segmentation by tiles
[x] Step 7: Compute intersection between segment and region
Group sampling:
[x] Step 8: Generate tile's envelope
[x] Step 9: Generate a region vector
[x] Step 10: Prepare samples
[x] Step 11: merge samples by models
[x] Step 12: Intersect samples and segmentation
[x] Step 13: Compute zonal statistics for learning polygons
Group learning:
[x] Step 14: learn model for obia
Group classification:
[x] Step 15: Classify tiles using zonal stats
Group validation:
[x] Step 16: Reassemble tiles and compute confusion matrix
[x] Step 17: Merge all vector tiles
[x] Step 18: Merge all metrics files
Once the processing starts, a large amount of information will be printed, most of it about the dask-scheduler.
Did it all go well?
iota2 has a logging system. Each step has its has its own log folder, available in the output_path/logs directory. The log directory is structured as:
.
├── check_inputs_classif_workflow
│ ├── check_inputs.err
│ └── check_inputs.out
├── CommonMasks
│ ├── common_mask_T31TCJ.err
│ └── common_mask_T31TCJ.out
├── compute_intersection_seg_regions
│ ├── compute_inter_sef_regs_T31TCJ.err
│ └── compute_inter_sef_regs_T31TCJ.out
├── compute_metrics_obia
│ ├── compute_metrics_T31TCJ.err
│ └── compute_metrics_T31TCJ.out
├── Envelope
│ ├── tiles_envelopes.err
│ └── tiles_envelopes.out
├── genRegionVector
│ ├── region_generation.err
│ └── region_generation.out
├── html
│ ├── configuration_file.html
│ ├── environment_info.html
│ ├── genindex.html
│ ├── index.html
│ ├── input_files_content.html
│ ├── objects.inv
│ ├── output_path_content.html
│ ├── s2_path_content.html
│ ├── search.html
│ ├── searchindex.js
│ ├── source
│ │ ├── check_inputs.out
│ │ ├── classif_tile_T31TCJ.out
│ │ ├── common_mask_T31TCJ.out
│ │ ├── compute_inter_sef_regs_T31TCJ.out
│ │ ├── compute_metrics_T31TCJ.out
│ │ ├── configuration_file.rst
│ │ ├── environment_info.rst
│ │ ├── index.rst
│ │ ├── input_files_content.rst
│ │ ├── intersect_T31TCJ.out
│ │ ├── merge_final_metrics0.out
│ │ ├── merge_model_1_seed_0.out
│ │ ├── merge_tile_0.out
│ │ ├── output_path_content.rst
│ │ ├── prepare_seg_T31TCJ.out
│ │ ├── preprocessing_T31TCJ.out
│ │ ├── region_generation.out
│ │ ├── s2_path_content.rst
│ │ ├── tasks_status_1.rst
│ │ ├── tasks_status_2.rst
│ │ ├── tiles_envelopes.out
│ │ ├── validity_raster_T31TCJ.out
│ │ ├── vector_form_T31TCJ.out
│ │ └── zonal_stats_learn_T31TCJ_seed_0.out
│ ├── _sources
│ │ ├── configuration_file.rst.txt
│ │ ├── environment_info.rst.txt
│ │ ├── index.rst.txt
│ │ ├── input_files_content.rst.txt
│ │ ├── output_path_content.rst.txt
│ │ ├── s2_path_content.rst.txt
│ │ ├── tasks_status_1.rst.txt
│ │ └── tasks_status_2.rst.txt
│ ├── _static
│ │ ├── basic.css
│ │ ├── css
│ │ │ ├── badge_only.css
│ │ │ ├── fonts
│ │ │ │ ├── fontawesome-webfont.eot
│ │ │ │ ├── fontawesome-webfont.svg
│ │ │ │ ├── fontawesome-webfont.ttf
│ │ │ │ ├── fontawesome-webfont.woff
│ │ │ │ ├── fontawesome-webfont.woff2
│ │ │ │ ├── lato-bold-italic.woff
│ │ │ │ ├── lato-bold-italic.woff2
│ │ │ │ ├── lato-bold.woff
│ │ │ │ ├── lato-bold.woff2
│ │ │ │ ├── lato-normal-italic.woff
│ │ │ │ ├── lato-normal-italic.woff2
│ │ │ │ ├── lato-normal.woff
│ │ │ │ ├── lato-normal.woff2
│ │ │ │ ├── Roboto-Slab-Bold.woff
│ │ │ │ ├── Roboto-Slab-Bold.woff2
│ │ │ │ ├── Roboto-Slab-Regular.woff
│ │ │ │ └── Roboto-Slab-Regular.woff2
│ │ │ └── theme.css
│ │ ├── doctools.js
│ │ ├── documentation_options.js
│ │ ├── file.png
│ │ ├── jquery-3.5.1.js
│ │ ├── jquery.js
│ │ ├── js
│ │ │ ├── badge_only.js
│ │ │ ├── html5shiv.min.js
│ │ │ ├── html5shiv-printshiv.min.js
│ │ │ └── theme.js
│ │ ├── language_data.js
│ │ ├── minus.png
│ │ ├── plus.png
│ │ ├── pygments.css
│ │ ├── searchtools.js
│ │ ├── underscore-1.3.1.js
│ │ └── underscore.js
│ ├── tasks_status_1.html
│ └── tasks_status_2.html
├── intersect_seg_learn
│ ├── intersect_T31TCJ.err
│ └── intersect_T31TCJ.out
├── learning_zonal_statistics
│ ├── zonal_stats_learn_T31TCJ_seed_0.err
│ └── zonal_stats_learn_T31TCJ_seed_0.out
├── merge_final_metrics
│ ├── merge_final_metrics0.err
│ └── merge_final_metrics0.out
├── merge_tiles_obia
│ ├── merge_tile_0.err
│ └── merge_tile_0.out
├── obia_classification
│ ├── classif_tile_T31TCJ.err
│ └── classif_tile_T31TCJ.out
├── obia_learning
│ ├── model_1_seed_0.err
│ └── model_1_seed_0.out
├── PixelValidity
│ ├── validity_raster_T31TCJ.err
│ └── validity_raster_T31TCJ.out
├── prepare_obia_seg
│ ├── prepare_seg_T31TCJ.err
│ └── prepare_seg_T31TCJ.out
├── run_informations.txt
├── samplesMerge
│ ├── merge_model_1_seed_0.err
│ └── merge_model_1_seed_0.out
├── sensorsPreprocess
│ ├── preprocessing_T31TCJ.err
│ └── preprocessing_T31TCJ.out
├── tasks_status_i2_obia_1.svg
├── tasks_status_i2_obia_2.svg
└── VectorFormatting
├── vector_form_T31TCJ.err
└── vector_form_T31TCJ.out
In these directories two kinds of log can be found *_out.log and *_err.log.
The errors are compiled in the “err” file and the standard output in the “out” file.
With the dask scheduler, iota2 gos as far as possible while the data required for the next steps is available.
To simplify the error identification, an interactive graph is produced in a html page.
To open it, open the index.html file in the html folder.
The nodes in the graph can have three colors (red: error, blue: done, orange: unprocessed).
By clicking on a graph node, the corresponding log file is opened.
If despite all this information, the errors can not be identified or solved, the iota2 developers can help. The simplest way to ask for help is to create an issue on framagit by adding the archive available in log directory.
Output tree structure
In this section, the iota2 outputs available after a proper run are described
.
├── classif
│ ├── classif_tmp
│ ├── MASK
│ │ └── MASK_region_1_T31TCJ.tif
│ ├── reduced
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.cpg
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.dbf
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.prj
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.shp
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.shx
│ ├── Vectorized_map_tile_T31TCJ_seed_0_clipped.dbf
│ ├── Vectorized_map_tile_T31TCJ_seed_0_clipped.prj
│ ├── Vectorized_map_tile_T31TCJ_seed_0_clipped.shp
│ ├── Vectorized_map_tile_T31TCJ_seed_0_clipped.shx
│ ├── Vectorized_map_tile_T31TCJ_seed_0_clipped.sqlite
│ ├── Vectorized_map_tile_T31TCJ_seed_0.cpg
│ ├── Vectorized_map_tile_T31TCJ_seed_0.dbf
│ ├── Vectorized_map_tile_T31TCJ_seed_0.prj
│ ├── Vectorized_map_tile_T31TCJ_seed_0.shp
│ ├── Vectorized_map_tile_T31TCJ_seed_0.shx
│ └── zonal_stats
│ ├── seg_T31TCJ_region_1_grid_*_seed_0.cpg
│ ├── seg_T31TCJ_region_1_grid_*_seed_0.csv
│ ├── seg_T31TCJ_region_1_grid_*_seed_0.dbf
│ ├── seg_T31TCJ_region_1_grid_*_seed_0.prj
│ ├── seg_T31TCJ_region_1_grid_*_seed_0.shp
│ ├── seg_T31TCJ_region_1_grid_*_seed_0.shx
│ ├── Sentinel2_T31TCJ_samples_seed_0_region_1_part_*_stats.xml
│ └── Stats_labels.txt
├── config_model
├── dataAppVal
│ ├── bymodels
│ ├── T31TCJ_seed_0_learn.dbf
│ ├── T31TCJ_seed_0_learn.prj
│ ├── T31TCJ_seed_0_learn.shp
│ ├── T31TCJ_seed_0_learn.shx
│ ├── T31TCJ_seed_0_learn.sqlite
│ ├── T31TCJ_seed_0_val.dbf
│ ├── T31TCJ_seed_0_val.prj
│ ├── T31TCJ_seed_0_val.shp
│ ├── T31TCJ_seed_0_val.shx
│ └── T31TCJ_seed_0_val.sqlite
├── dataRegion
├── envelope
│ ├── T31TCJ.dbf
│ ├── T31TCJ.prj
│ ├── T31TCJ.shp
│ └── T31TCJ.shx
├── features
│ └── T31TCJ
│ ├── CloudThreshold_0.dbf
│ ├── CloudThreshold_0.prj
│ ├── CloudThreshold_0.shp
│ ├── CloudThreshold_0.shx
│ ├── nbView.tif
│ └── tmp
│ ├── MaskCommunSL.dbf
│ ├── MaskCommunSL.prj
│ ├── MaskCommunSL.shp
│ ├── MaskCommunSL.shx
│ ├── MaskCommunSL.tif
│ ├── Sentinel2_T31TCJ_input_dates.txt
│ ├── Sentinel2_T31TCJ_interpolation_dates.txt
│ └── Sentinel2_T31TCJ_reference.tif
├── final
│ ├── Classif_Seed_0.sqlite
│ ├── Confusion_Matrix_Classif_Seed_0.png
│ ├── RESULTS.txt
│ └── TMP
│ ├── Classif_Seed_0.csv
│ └── T31TCJ_confusion_matrix_seed_0.csv
├── formattingVectors
│ ├── T31TCJ
│ ├── T31TCJ.cpg
│ ├── T31TCJ.dbf
│ ├── T31TCJ.prj
│ ├── T31TCJ.shp
│ └── T31TCJ.shx
├── IOTA2_tasks_status.txt
├── learningSamples
│ ├── seg_T31TCJ_region_1_grid_*_seed_0_seed_0.cpg
│ ├── seg_T31TCJ_region_1_grid_*_seed_0_seed_0.csv
│ ├── seg_T31TCJ_region_1_grid_*_seed_0_seed_0.dbf
│ ├── seg_T31TCJ_region_1_grid_*_seed_0_seed_0.prj
│ ├── seg_T31TCJ_region_1_grid_*_seed_0_seed_0.shp
│ ├── seg_T31TCJ_region_1_grid_*_seed_0_seed_0.shx
│ ├── Stats_labels.txt
│ └── zonal_stats
│ ├── Sentinel2_T31TCJ_samples_seed_0_region_1_part_*_stats.xml
├── logs
│ ├── *
├── logs.zip
├── model
│ └── model_region_1_seed_0.txt
├── MyRegion.dbf
├── MyRegion.prj
├── MyRegion.shp
├── MyRegion.shx
├── reference_data.dbf
├── reference_data.prj
├── reference_data.shp
├── reference_data.shx
├── samplesSelection
│ ├── samples_region_1_seed_0.dbf
│ ├── samples_region_1_seed_0.prj
│ ├── samples_region_1_seed_0.shp
│ └── samples_region_1_seed_0.shx
├── segmentation
│ ├── grid_split
│ │ └── T31TCJ
│ │ ├── seg_T31TCJ_grid.cpg
│ │ ├── seg_T31TCJ_grid.dbf
│ │ ├── seg_T31TCJ_grid.shp
│ │ ├── seg_T31TCJ_grid.shx
│ │ ├── seg_T31TCJ_region_1_grid_*.cpg
│ │ ├── seg_T31TCJ_region_1_grid_*.dbf
│ │ ├── seg_T31TCJ_region_1_grid_*_mask.tif
│ │ ├── seg_T31TCJ_region_1_grid_*.prj
│ │ ├── seg_T31TCJ_region_1_grid_*.shp
│ │ ├── seg_T31TCJ_region_1_grid_*.shx
│ │ ├── seg_T31TCJ_region_1_grid_*.tif
│ ├── grid_split_learn
│ │ └── T31TCJ
│ │ ├── seg_T31TCJ_grid.cpg
│ │ ├── seg_T31TCJ_grid.dbf
│ │ ├── seg_T31TCJ_grid.shp
│ │ ├── seg_T31TCJ_grid.shx
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.cpg
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.dbf
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0_mask.tif
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.prj
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.shp
│ │ ├── seg_T31TCJ_region_1_grid_*_seed_0.shx
│ │ ├── seg_T31T CJ_region_1_grid_*_seed_0.tif
│ ├── learning_samples_T31TCJ_seed_0.cpg
│ ├── learning_samples_T31TCJ_seed_0.dbf
│ ├── learning_samples_T31TCJ_seed_0.prj
│ ├── learning_samples_T31TCJ_seed_0.shp
│ ├── learning_samples_T31TCJ_seed_0.shx
│ ├── seg_T31TCJ.cpg
│ ├── seg_T31TCJ.dbf
│ ├── seg_T31TCJ.prj
│ ├── seg_T31TCJ.shp
│ ├── seg_T31TCJ.shx
│ └── tmp
│ ├── grid_intersect_seg_T31TCJ.cpg
│ ├── grid_intersect_seg_T31TCJ.dbf
│ ├── grid_intersect_seg_T31TCJ.prj
│ ├── grid_intersect_seg_T31TCJ_seed_0.cpg
│ ├── grid_intersect_seg_T31TCJ_seed_0.dbf
│ ├── grid_intersect_seg_T31TCJ_seed_0.prj
│ ├── grid_intersect_seg_T31TCJ_seed_0.shp
│ ├── grid_intersect_seg_T31TCJ_seed_0.shx
│ ├── grid_intersect_seg_T31TCJ.shp
│ ├── grid_intersect_seg_T31TCJ.shx
│ ├── intersect_T31TCJ_learning_samples.cpg
│ ├── intersect_T31TCJ_learning_samples.dbf
│ ├── intersect_T31TCJ_learning_samples.prj
│ ├── intersect_T31TCJ_learning_samples.shp
│ ├── intersect_T31TCJ_learning_samples.shx
│ ├── regions_T31TCJ.cpg
│ ├── regions_T31TCJ.dbf
│ ├── regions_T31TCJ.prj
│ ├── regions_T31TCJ.shp
│ ├── regions_T31TCJ.shx
│ ├── seg_T31TCJ.dbf
│ ├── seg_T31TCJ.prj
│ ├── seg_T31TCJ.shp
│ ├── seg_T31TCJ.shx
│ ├── seg_T31TCJ.tif
│ ├── T31TCJ_samples_without_region.cpg
│ ├── T31TCJ_samples_without_region.dbf
│ ├── T31TCJ_samples_without_region.prj
│ ├── T31TCJ_samples_without_region.shp
│ └── T31TCJ_samples_without_region.shx
├── shapeRegion
│ ├── MyRegion_region_1_T31TCJ.dbf
│ ├── MyRegion_region_1_T31TCJ.prj
│ ├── MyRegion_region_1_T31TCJ.shp
│ ├── MyRegion_region_1_T31TCJ.shx
│ └── MyRegion_region_1_T31TCJ.tif
└── stats
classif
Temporary classification maps, for each tile and region.
The folder zonal_stats contains all shapefiles containing features, the corresponding features are provided by the xml files according to the part ID. Stats_labels.txt contains all features names used for training and classification.
Folder reduced contains all shapefiles containing classification from zonal_stats folder. All the features are removed in these files.
Vectorized_map_tile_XXXX_seed_N.shp is the merge of all parts for each tile XXXX.
Vectorized_map_tile_XXXX_seed_N_clipped.sqlite is the classification for the tile XXXX, clipped with the corresponding tile enveloppe.
config_model
Empty
dataRegion
When using eco-climatic regions, it contains the vector data split by region.
envelope
Contains shapefiles, one for each tile. Used to ensure tile priority, with no overlap.
formattingVectors
The learning samples contained in each tile. Shapefiles in which pixel values from time series have been extracted.
samplesSelection
Shapefiles containing all polygons selected for the training stage.
stats
Optional xml statistics to standardize the data before learning (svm…).
dataAppVal
Shapefiles obtained after spliting reference data between learning and validation set according a ratio.
final
This folder contains the final products of iota2.
learningSamples
Shapefile containing learning samples by region.
model
The learned models
shapeRegion
Shapefiles indicating intersection between tiles and regions.
features
For each tile, contains useful information:
nbView.tif: number of times a pixel is seen in the whole time series (i.e., excluding clouds, shadows, staturation and no-data)CloudThreshold.shp: this database is used to mask training polygons according to a number of clear dates. See cloud_threshold <i2_classification_builder.rst> parameter.tmp/MaskComminSL.*: the common scene of all sensors for this tile.tmp/Sentinel2L3A_T31TCJ_reference.tif: the image, generated by iota2, used for reprojecting data.tmp/Sentinel2L3A_T31TCJ_input_dates.txt: the list of dates detected ins2_pathfor the current tile.
segmentation
tmp: folder which contains working files, like the segmentation clipped on tile, the tiles grids, …grid_split_learn: folder with shapefiles resulting from intersection between learning samples and the segmentation by tile, divided along the grid in several parts.grid_split: folder with shapefiles resulting from intersection between regions and the segmentation by tile, divided along the grid in several parts.
learning_samples_XXXX_seed_0.shp contains the intersection between learning shapefile and the segmentation.
seg_XXXX.shp: the segmentation where each segment is asociated to a region.
Final products
All final products will be generated in the final directory.
OBIA outputs are stored in sqlite format.
Land cover map
Your Classif_Seed_0.sqlite should look like this one (after loading the style file):
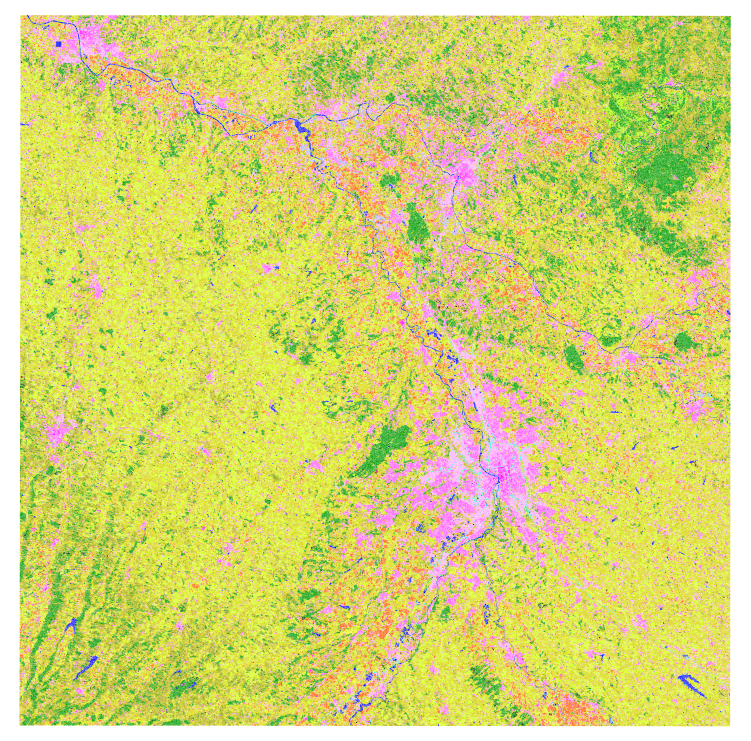
Classif_Seed_0.sqlite Example
The classification results are contained in two columns: data_field (the name change according to the configuration parameter data_field) and confidence if the classifier provides confidence.
The OTB vector classification application does not return probabilities.
This tutorial uses a very small dataset, so the results are not really good. The simplest method to get better results can consist in using a longer time series, improving the reference data for training, use more significant statistics, …
Measuring quality
The OBIA workflow provides two statistical results:
a text file
#row = reference
#col = production
*********** Matrice de confusion ***********
| batis denses | batis diffus | zones ind et com | surfaces routes | colza | cereales à pailles| protéagineux | soja | tournesol | maïs | tubercules/racines| prairies | vergers | vignes | forets de feuillus|forets de coniferes| pelouses | landes ligneuses | eau
batis denses | 141.00 | 1378.00 | 864.00 | 50.00 | 0.00 | 64.00 | 0.00 | 5.00 | 201.00 | 30.00 | 0.00 | 30.00 | 0.00 | 0.00 | 2.00 | 0.00 | 1.00 | 0.00 | 6.00 | batis denses
batis diffus | 52.00 | 2964.00 | 1905.00 | 28.00 | 0.00 | 62.00 | 0.00 | 1.00 | 615.00 | 36.00 | 0.00 | 162.00 | 1.00 | 3.00 | 35.00 | 4.00 | 9.00 | 0.00 | 5.00 | batis diffus
zones ind et com | 45.00 | 1762.00 | 5601.00 | 230.00 | 0.00 | 436.00 | 0.00 | 23.00 | 1894.00 | 186.00 | 0.00 | 476.00 | 26.00 | 7.00 | 139.00 | 3.00 | 20.00 | 0.00 | 23.00 | zones ind et com
surfaces routes | 0.00 | 30.00 | 258.00 | 349.00 | 0.00 | 17.00 | 0.00 | 10.00 | 382.00 | 9.00 | 0.00 | 41.00 | 6.00 | 1.00 | 12.00 | 1.00 | 1.00 | 0.00 | 3.00 | surfaces routes
colza | 2.00 | 141.00 | 341.00 | 8.00 | 41.00 | 376.00 | 0.00 | 15.00 | 6485.00 | 59.00 | 0.00 | 278.00 | 7.00 | 3.00 | 239.00 | 2.00 | 6.00 | 0.00 | 0.00 | colza
cereales à pailles| 20.00 | 1044.00 | 1171.00 | 14.00 | 2.00 | 8790.00 | 0.00 | 66.00 | 3622.00 | 1182.00 | 0.00 | 598.00 | 6.00 | 10.00 | 178.00 | 1.00 | 28.00 | 0.00 | 2.00 | cereales à pailles
protéagineux | 1.00 | 69.00 | 65.00 | 1.00 | 0.00 | 163.00 | 0.00 | 4.00 | 935.00 | 17.00 | 0.00 | 29.00 | 1.00 | 0.00 | 20.00 | 1.00 | 1.00 | 0.00 | 2.00 | protéagineux
soja | 13.00 | 635.00 | 861.00 | 9.00 | 9.00 | 3105.00 | 0.00 | 222.00 | 8037.00 | 916.00 | 0.00 | 560.00 | 14.00 | 6.00 | 242.00 | 8.00 | 6.00 | 0.00 | 13.00 | soja
tournesol | 6.00 | 1542.00 | 2754.00 | 27.00 | 55.00 | 4551.00 | 0.00 | 201.00 | 40687.00 | 748.00 | 0.00 | 2353.00 | 49.00 | 28.00 | 1171.00 | 23.00 | 58.00 | 0.00 | 12.00 | tournesol
maïs | 46.00 | 876.00 | 1168.00 | 22.00 | 50.00 | 3315.00 | 0.00 | 57.00 | 8664.00 | 2413.00 | 0.00 | 442.00 | 9.00 | 8.00 | 206.00 | 13.00 | 12.00 | 0.00 | 8.00 | maïs
tubercules/racines| 0.00 | 27.00 | 47.00 | 3.00 | 0.00 | 148.00 | 0.00 | 1.00 | 566.00 | 23.00 | 0.00 | 13.00 | 1.00 | 1.00 | 4.00 | 0.00 | 0.00 | 0.00 | 0.00 | tubercules/racines
prairies | 0.00 | 79.00 | 235.00 | 1.00 | 0.00 | 94.00 | 0.00 | 2.00 | 960.00 | 19.00 | 0.00 | 2542.00 | 2.00 | 8.00 | 392.00 | 1.00 | 85.00 | 0.00 | 1.00 | prairies
vergers | 0.00 | 49.00 | 315.00 | 5.00 | 0.00 | 39.00 | 0.00 | 2.00 | 263.00 | 8.00 | 0.00 | 115.00 | 287.00 | 1.00 | 36.00 | 0.00 | 0.00 | 0.00 | 3.00 | vergers
vignes | 5.00 | 173.00 | 134.00 | 0.00 | 0.00 | 78.00 | 0.00 | 1.00 | 260.00 | 12.00 | 0.00 | 49.00 | 2.00 | 42.00 | 0.00 | 0.00 | 6.00 | 0.00 | 0.00 | vignes
forets de feuillus| 0.00 | 17.00 | 55.00 | 0.00 | 1.00 | 5.00 | 0.00 | 0.00 | 386.00 | 2.00 | 0.00 | 474.00 | 1.00 | 0.00 | 1227.00 | 5.00 | 29.00 | 0.00 | 4.00 | forets de feuillus
forets de coniferes| 0.00 | 3.00 | 6.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 148.00 | 0.00 | 0.00 | 38.00 | 0.00 | 0.00 | 70.00 | 162.00 | 5.00 | 1.00 | 0.00 |forets de coniferes
pelouses | 0.00 | 13.00 | 61.00 | 0.00 | 0.00 | 29.00 | 0.00 | 3.00 | 210.00 | 1.00 | 0.00 | 176.00 | 0.00 | 1.00 | 54.00 | 16.00 | 21.00 | 0.00 | 9.00 | pelouses
landes ligneuses | 0.00 | 14.00 | 65.00 | 0.00 | 0.00 | 7.00 | 0.00 | 0.00 | 109.00 | 1.00 | 0.00 | 302.00 | 0.00 | 2.00 | 42.00 | 1.00 | 72.00 | 0.00 | 5.00 | landes ligneuses
eau | 1.00 | 2.00 | 34.00 | 48.00 | 0.00 | 0.00 | 0.00 | 14.00 | 65.00 | 12.00 | 0.00 | 3.00 | 0.00 | 0.00 | 0.00 | 8.00 | 0.00 | 0.00 | 2291.00 | eau
KAPPA : 0.308
OA : 0.463
Classes | Precision mean | Rappel mean | F-score mean | Confusion max
------------------------------------------------------------------------------------------------------------
batis denses | 0.425 | 0.051 | 0.091 | batis diffus, zones ind et com, tournesol
batis diffus | 0.274 | 0.504 | 0.355 | batis diffus, zones ind et com, tournesol
zones ind et com | 0.351 | 0.515 | 0.418 | zones ind et com, tournesol, batis diffus
surfaces routes | 0.439 | 0.312 | 0.364 | tournesol, surfaces routes, zones ind et com
colza | 0.259 | 0.005 | 0.010 | tournesol, cereales à pailles, zones ind et com
cereales à pailles | 0.413 | 0.525 | 0.462 | cereales à pailles, tournesol, maïs
protéagineux | 0.000 | 0.000 | 0.000 | tournesol, cereales à pailles, batis diffus
soja | 0.353 | 0.015 | 0.029 | tournesol, cereales à pailles, maïs
tournesol | 0.546 | 0.750 | 0.632 | tournesol, cereales à pailles, zones ind et com
maïs | 0.425 | 0.139 | 0.210 | tournesol, cereales à pailles, maïs
tubercules/racines | 0.000 | 0.000 | 0.000 | tournesol, cereales à pailles, zones ind et com
prairies | 0.293 | 0.575 | 0.388 | prairies, tournesol, forets de feuillus
vergers | 0.697 | 0.256 | 0.374 | zones ind et com, vergers, tournesol
vignes | 0.347 | 0.055 | 0.095 | tournesol, batis diffus, zones ind et com
forets de feuillus | 0.302 | 0.556 | 0.391 | forets de feuillus, prairies, tournesol
forets de coniferes | 0.651 | 0.372 | 0.474 | forets de coniferes, tournesol, forets de feuillus
pelouses | 0.058 | 0.035 | 0.044 | tournesol, prairies, zones ind et com
landes ligneuses | 0.000 | 0.000 | 0.000 | prairies, tournesol, pelouses
eau | 0.960 | 0.925 | 0.942 | eau, tournesol, surfaces routes
an image of the confusion matrix
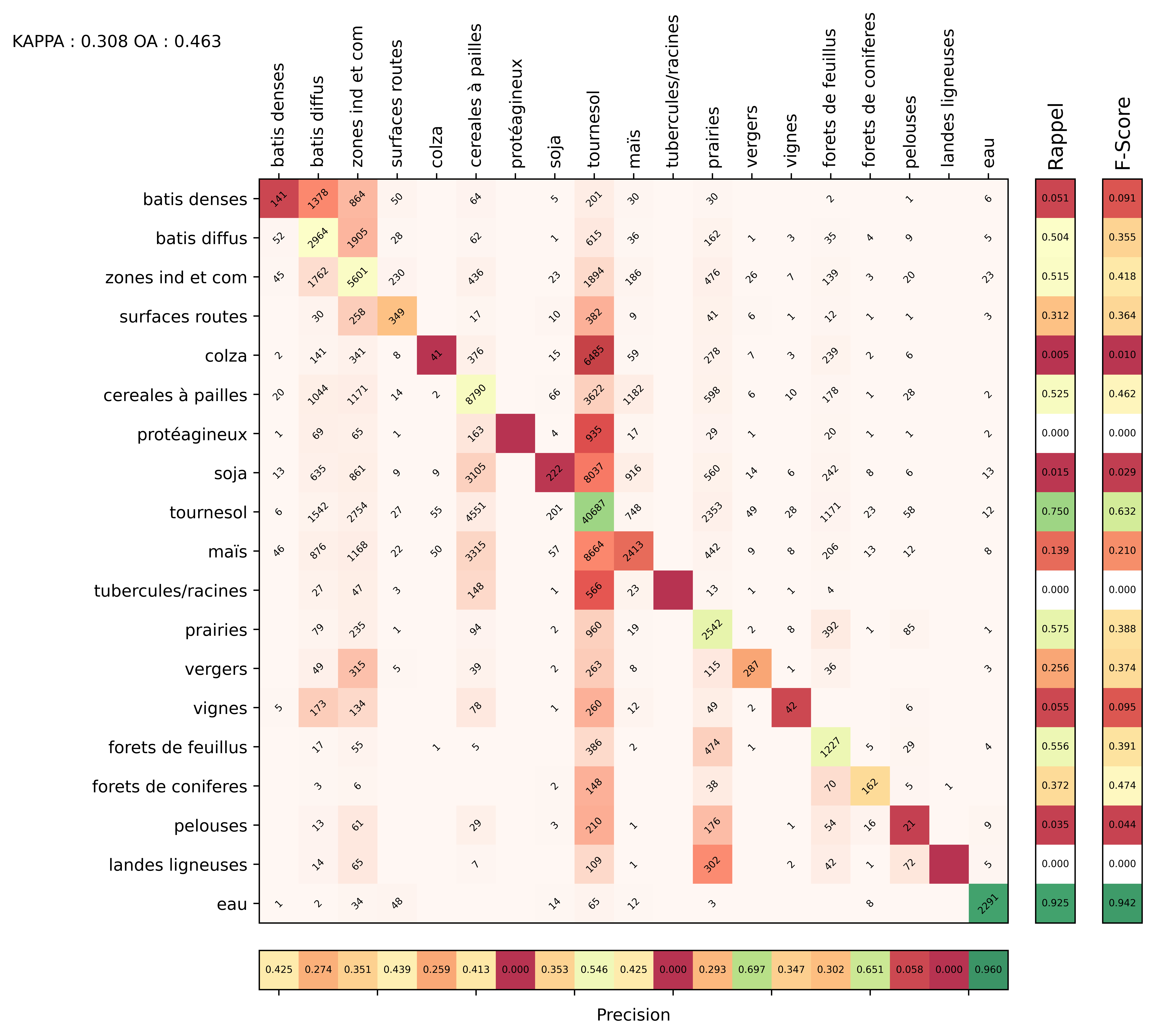
Example of confusion matrix