Iota2 regression
While iota2 was initially designed for land cover classification (prediction of a discrete variable), it also allows to perform regression (prediction of a continuous variable). This chapter introduces the use of iota2 regression builder.
The regression pipeline is divided in the following groups:
Init: pre-processing steps;
Sampling: input data is split between train and validation set, training pixels are selected and their corresponding features extracted;
Training: target values are standardized (mean to 0 and std to 1) and passed to a model for training; Features can be also standardized;
Prediction: the trained model is used to predict pixel values on a map (tile-wise processing);
Mosaic: tiles are merged together;
Validation: prediction is evaluated with the validation samples.
In regression mode, two learn/predict workflows are available:
Pytorch: neural networks built inside
iota2;scikit-learn: multiple models available from this library.
Configuration file
iota2 is configured through several parameters, some of them are specfic to iota2 and some belong to other libraries such as scikit-learn.
These parameters allow to select the operations to be carried out and their parameters. A documentation of all these parameters is provided here. The user defines these paramereters in a configuration file (a human readable text file) that is read by iota2 at upon start. The file is structured into sections, each section containing several fields.
Below, two sample configuration files for performing regression, one for each workflow are shown.
Pytorch configuration template
chain:
{
output_path: "/results_tuto"
s2_path: "/IOTA2_TUTO_REGRESSION/sensor_data"
ground_truth: "/IOTA2_TUTO_REGRESSION/vector_data/reference_ndvi.shp"
remove_output_path: True
list_tile: "T31TCJ T31TDJ"
data_field: "ndvi"
spatial_resolution: 10
proj: "EPSG:2154"
first_step: "init"
last_step: "validation"
}
arg_train:
{
runs: 1
sample_selection:
{
"sampler": "periodic",
"strategy": "all",
}
deep_learning_parameters:
{
dl_name: "MLPRegressor"
epochs: 10
model_optimization_criterion: "MSE"
model_selection_criterion: "MSE"
hyperparameters_solver: {"batch_size": [1000], "learning_rate": [0.001]}
}
}
python_data_managing:
{
number_of_chunks: 10
}
builders:
{
builders_class_name: ["I2Regression"]
}
scikit-learn configuration template
chain:
{
output_path: "/results_tuto"
s2_path: "/IOTA2_TUTO_REGRESSION/sensor_data"
ground_truth: "/IOTA2_TUTO_REGRESSION/vector_data/reference_ndvi.shp"
remove_output_path: True
list_tile: "T31TCJ T31TDJ"
data_field: "ndvi"
spatial_resolution: 10
proj: "EPSG:2154"
first_step: "init"
last_step: "validation"
}
arg_train:
{
runs: 3
sample_selection:
{
"sampler": "periodic"
"strategy": "all"
}
}
scikit_models_parameters:
{
model_type: "HuberRegressor"
standardization: True
keyword_arguments:
{
epsilon: 1.35
}
}
python_data_managing:
{
number_of_chunks: 10
}
builders:
{
builders_class_name: ["I2Regression"]
}
multi_run_fusion:
{
merge_run: True
merge_run_method: 'mean'
}
Configuration template explanation
To use the regression mode, the user must provide a builders section with only I2Regression. This builder is not compatible with other builders.
builders:
{
builders_class_name: [I2Regression"]
}
The data_field parameter of the ground_truth file represents the target value used to train de model. It must be lowercase, without underscores, and point to an integer or float field of your ground_truth file.
The ground_truth parameter must be a shape file containing polygons or points
If you want to use the Pytorch workflow, fill in the deep_learning_parameters field of the arg_train section.
If you want to use the scikit-learn workflow, fill in the scikit_models_parameters section.
The two are not compatible. One and only one of these must be set.
In both cases, you must provide the python_data_managing / number_of_chunks field to split the tile prediction in chunks. A too low value will result in larger chunks and may not fit in memory while a too high value will increase the scheduler overhead.
python_data_managing:
{
number_of_chunks: 20
}
Pytorch workflow
For full configuration, see Deep Learning in iota2.
To perform regression with Pytorch, you must set dl_name to one of the available models for regression:
MLPRegressormulti layer perceptron defined in torch_nn_bank.pyuser defined model in
dl_moduleinheriting fromIota2NeuralNetwork
arg_train:
{
deep_learning_parameters:
{
dl_name: "MLPRegressor" # <-- here
model_optimization_criterion: "MSE"
model_selection_criterion: "MSE"
# ...
}
}
The regression mode offers multiple criteria.
Available options for model_optimization_criterion:
After training models for all combinations of hyperparameters in hyperparameters_solver, the best model will be selected according to the model_selection_criterion which accepts the same values.
The prediction will then be made by chunks as configured in python_data_managing.
Scikit-learn workflow
For full configuration, see iota2 and scikit-learn machine learning algorithms.
To perform regression with scikit-learn, you must set model_type to one of the available models for regression:
scikit_models_parameters:
{
model_type: "RandomForestRegressor"
standardization: True
keyword_arguments:
{
criterion: "squared_error"
}
}
Additional parameters can be given to the model using the keyword_arguments field.
The prediction will then be made by chunks as configured in python_data_managing.
Fusion between multiple runs
If you perform several executions (runs argument from section arg_train> 1), the results of the different executions can be merged by setting to True the parameter merge_run of the multi_run_fusion section.
The merge itself can be done as the mean or the median value of the predicted pixel values over the different runs. The method choice must be indicated with the parameter merge_run_method in the section multi_run_fusion
multi_run_fusion:
{
merge_run: True
merge_run_method: 'mean'
}
Run iota2
For complete documentation about iota2 command line arguments, see Going further with iota2 launching parameters.
You can then run iota2 using the following command line to validate your configuration file.
Iota2.py -config config_regression.cfg -only_summary
In scikit-learn mode, it should produce something similar to:
Group init:
[x] Step 1: Sensors pre-processing
[x] Step 2: Generate a common masks for each sensors
[x] Step 3: Compute validity raster by tile
Group sampling:
[x] Step 4: Generate tile's envelope
[x] Step 5: Generate a region vector
[x] Step 6: Prepare samples
[x] Step 7: merge samples by models
[x] Step 8: Generate samples statistics by models
[x] Step 9: Select pixels in learning polygons by models
[x] Step 10: Split pixels selected to learn models by tiles
[x] Step 11: Extract pixels values by tiles
[x] Step 12: Merge samples dedicated to the same model
Group training:
[x] Step 13: Train scikit random forest
Group prediction:
[x] Step 14: Predict with scikit random forest
[x] Step 15: Merge tile's classification's part
Group mosaic:
[x] Step 16: Mosaic
[x] Step 17: Merge final regressions
Group validation:
[x] Step 18: Generate regression metrics
[x] Step 19: Generate regression metrics for multi tile and multi run configurations
[x] Step 20: Merge final validation
Inspect output
After iota2 has ended, you can go to the output folder and see your results in the final folder (see Final folder). If the results are not there, you should find why in the logs. You can look at the graphs tasks_status_i2_regression_<i>.svg to find out which step has failed. Then you can get more details about the error in the logs or by re-launching the given step in debug mode (-starting_step <j> -ending_step <j> -scheduler_type debug).
Output tree
This output tree is based on a “scikit-learn” run. Yours can differ slightly if you choose different parameters.
- /Output_Regression
- output folder
output folder defined in config file output_path
- ! classif
- per tile prediction maps
- Contains regression maps, for each tile and each region. They will be merged in the
finaldirectory.
Regression_T31TCJ_model_1_seed_0.tif
Regression_T31TDJ_model_1_seed_0.tif
- ! MASK
MASK_region_1_T31TCJ.tif
MASK_region_1_T31TDJ.tif
- ! config_model
(empty)
- ! dataAppVal
- split train / validation samples
- Shapefiles obtained after spliting reference data between learning and validation set according a ratio.
- ! bymodels
(empty)
- T31TCJ_seed_0_learn.sqlite
learning polygons
- T31TCJ_seed_0_val.sqlite
validation polygons
- T31TCJ_seed_0_val.xml
validation statistics for sample extraction
- T31TCJ_seed_0_val_point.sqlite
points sampled in validation polygons
- T31TCJ_seed_0_val_predicted_Regression_Seed_0.sqlite
- predicted values
- these values are pixels extracted from the final mosaic map at the location of validation data
- T31TDJ_seed_0_learn.sqlite
learning polygons
- T31TDJ_seed_0_val.sqlite
validation polygons
- T31TDJ_seed_0_val.xml
validation statistics for sample extraction
- T31TDJ_seed_0_val_point.sqlite
points sampled in validation polygons
- T31TDJ_seed_0_val_predicted_Regression_Seed_0.sqlite
- predicted values
- these values are pixels extracted from the final mosaic map at the location of validation data
- fusion_val_Regression_Seed_0.sqlite
validation polygons of the mosaic
- ! dataRegion
- vector data split by region
- When using eco-climatic region, contains the vector data split by region.
(empty)
- ! envelope
- shapefiles
- Contains shapefiles, one for each tile.Used to ensure tile priority, with no overlap.
T31TDJ.dbf
T31TCJ.prj
T31TCJ.shp
T31TCJ.shx
T31TDJ.dbf
T31TDJ.prj
T31TDJ.shp
T31TDJ.shx
- ! features
- useful information
- for each tile, contains useful information
- T31TCJ
- ! tmp
- temporary folder
- folder created temporarily during the chain execution
MaskCommunSL.dbf
MaskCommunSL.prj
- MaskCommunSL.shp
- common scene
- the common scene of all sensors for this tile.
MaskCommunSL.shx
MaskCommunSL.tif
- Sentinel2L3A_T31TCJ_reference.tif
- reference image
- the image, generated by iota2, used for reprojecting data
- Sentinel2L3A_T31TCJ_input_dates.txt
- list of dates
- the list of date detected in
s2_pathfor the current tile.
Sentinel2_T31TCJ_interpolation_dates.txt
CloudThreshold_0.dbf
CloudThreshold_0.prj
- CloudThreshold_0.shp
- database used as mask
- This database is used to mask training polygons according to a number of clear date. See cloud_threshold parameter
CloudThreshold_0.shx
- nbView.tif
- number visits
- number of time a pixel is seen in the whole time series (i.e., excluding clouds, shadows, staturation and no-data)
- T31TDJ
- ! tmp
- temporary folder
- folder created temporarily during the chain execution
MaskCommunSL.dbf
MaskCommunSL.prj
- MaskCommunSL.shp
- common scene
- the common scene of all sensors for this tile.
MaskCommunSL.shx
MaskCommunSL.tif
- Sentinel2L3A_T31TDJ_reference.tif
- reference image
- the image, generated by iota2, used for reprojecting data
- Sentinel2L3A_T31TDJ_input_dates.txt
- list of dates
- the list of date detected in
s2_pathfor the current tile.
Sentinel2_T31TDJ_interpolation_dates.txt
CloudThreshold_0.dbf
CloudThreshold_0.prj
- CloudThreshold_0.shp
- database used as mask
- This database is used to mask training polygons according to a number of clear date. See cloud_threshold parameter
CloudThreshold_0.shx
- nbView.tif
- number visits
- number of time a pixel is seen in the whole time series (i.e., excluding clouds, shadows, staturation and no-data)
- final
- final producs
- This folder contains the final products of iota2.All final products will be generated in the
finaldirectorysee Final folder for details
- ! merge_final_classification
validation files for the evaluation of the fusion map * (empty)
- ! TMP
T31TCJ_seed_0.tif
T31TDJ_seed_0.tif
- Regression_Seed_0.tif
- prediction map
- destandardized float32 values predicted by the regression model
- T31TCJ_metrics.csv
- summary of metrics
- metrics per tile summarized over multiple seeds
- T31TCJ_seed0_metrics.csv
- metrics per seed
- metrics (mae, mse…) between predicted and actual values
- T31TDJ_metrics.csv
- summary of metrics
- metrics per tile summarized over multiple seeds
- T31TDJ_seed0_metrics.csv
- metrics per seed
- metrics (mae, mse…) between predicted and actual values
- mosaic_seed0_metrics.csv
metrics over the mosaic
- ! formattingVectors
- learning samples
- The learning samples contained in each tiles.Shapefiles in which pixel values from time series have been extracted.
- ! T31TCJ
- temporary directory
- This is a temporary working directory, intermediate files are (re)moved after step completion.
(empty)
T31TCJ.cpg
T31TCJ.dbf
T31TCJ.prj
T31TCJ.shp
T31TCJ.shx
- ! T31TDJ
- temporary directory
- This is a temporary working directory, intermediate files are (re)moved after step completion.
(empty)
T31TDJ.cpg
T31TDJ.dbf
T31TDJ.prj
T31TDJ.shp
T31TDJ.shx
- ! learningSamples
- learning samples
- Sqlite file containing learning samples by regions.Also contains a CSV file containing statistics about samples balance for each seed. See tracing back samples to generate this file manually.
class_statistics_seed0_learn.csv
Samples_region_1_seed0_learn.sqlite
T31TCJ_region_1_seed0_Samples_learn.sqlite
T31TDJ_region_1_seed0_Samples_learn.sqlite
- logs
- logs
- output logs of iota2here sorted by creation order (not alphabetic)
- ! SensorsPreprocess
preprocessing_T31TCJ.err
preprocessing_T31TCJ.out
preprocessing_T31TDJ.err
preprocessing_T31TDJ.out
- ! CommonMasks
common_mask_T31TCJ.err
common_mask_T31TCJ.out
common_mask_T31TDJ.err
common_mask_T31TDJ.out
- ! PixelValidity
validity_raster_T31TCJ.err
validity_raster_T31TCJ.out
validity_raster_T31TDJ.err
validity_raster_T31TDJ.out
- ! Envelope
tiles_envelopes.err
tiles_envelopes.out
- ! GenRegionVector
region_generation.err
region_generation.out
- ! VectorFormatting
vector_form_T31TCJ.err
vector_form_T31TCJ.out
vector_form_T31TDJ.err
vector_form_T31TDJ.out
- ! SamplesMerge
merge_model_1_seed_0.err
merge_model_1_seed_0.out
- ! StatsSamplesModel
stats_1_S_0_T_T31TCJ.err
stats_1_S_0_T_T31TCJ.out
stats_1_S_0_T_T31TDJ.err
stats_1_S_0_T_T31TDJ.out
- ! SamplingLearningPolygons
s_sel_model_1_seed_0.err
s_sel_model_1_seed_0.out
- ! SamplesByTiles
merge_samples_T31TCJ.err
merge_samples_T31TCJ.out
merge_samples_T31TDJ.err
merge_samples_T31TDJ.out
- ! SamplesExtraction
extraction_T31TCJ.err
extraction_T31TCJ.out
extraction_T31TDJ.err
extraction_T31TDJ.out
- ! SamplesByModels
merge_model_1_seed_0_usually.err
merge_model_1_seed_0_usually.out
- ! TrainRegressionScikit
learning_model_1_seed_0_hyp_0.err
learning_model_1_seed_0_hyp_0.out
- ! PredictRegressionScikit
regression_T31TCJ_model_1_seed_0_0.err
regression_T31TCJ_model_1_seed_0_0.out
regression_T31TCJ_model_1_seed_0_1.err
regression_T31TCJ_model_1_seed_0_1.out
regression_T31TCJ_model_1_seed_0_2.err
regression_T31TCJ_model_1_seed_0_2.out
regression_T31TDJ_model_1_seed_0_0.err
regression_T31TDJ_model_1_seed_0_0.out
regression_T31TDJ_model_1_seed_0_1.err
regression_T31TDJ_model_1_seed_0_1.out
regression_T31TDJ_model_1_seed_0_2.err
regression_T31TDJ_model_1_seed_0_2.out
- ! ScikitClassificationsMerge
classif_T31TCJ_model_1_seed_0_mosaic.err
classif_T31TCJ_model_1_seed_0_mosaic.out
classif_T31TDJ_model_1_seed_0_mosaic.err
classif_T31TDJ_model_1_seed_0_mosaic.out
- ! Mosaic
mosaic.err
mosaic.out
- ! GenerateRegressionMetrics
regression_metrics_T31TCJ_seed_0.err
regression_metrics_T31TCJ_seed_0.out
regression_metrics_T31TDJ_seed_0.err
regression_metrics_T31TDJ_seed_0.out
- ! GenerateRegressionMetricsSummary
regression_metrics_mosaic_seed_0.err
regression_metrics_mosaic_seed_0.out
tasks_status_i2_regression_1.svg
tasks_status_i2_regression_2.svg
tasks_status_i2_regression_3.svg
- run_information.txt
- summary
- summary as displayed by “only_summary” option
- ! model
- desc
- The learned models
model_1_seed_0.txt
- ! samplesSelection
- shapefiles
- Shapefiles containing points (or pixels coordinates) selected for training stage.Also contains a CSV summary of the actual number of samples per class
samples_region_1_seed_0.dbf
samples_region_1_seed_0_outrates.csv
samples_region_1_seed_0.prj
samples_region_1_seed_0_selection.sqlite
samples_region_1_seed_0.shp
samples_region_1_seed_0.shx
samples_region_1_seed_0.xml
T31TCJ_region_1_seed_0_stats.xml
T31TCJ_samples_region_1_seed_0_selection.sqlite
T31TCJ_selection_merge.sqlite
T31TDJ_region_1_seed_0_stats.xml
T31TDJ_samples_region_1_seed_0_selection.sqlite
T31TDJ_selection_merge.sqlite
- ! shapeRegion
- desc
- Shapefiles indicating intersection between tiles and region.
MyRegion_region_1_T31TCJ.dbf
MyRegion_region_1_T31TCJ.prj
MyRegion_region_1_T31TCJ.shp
MyRegion_region_1_T31TCJ.shx
MyRegion_region_1_T31TCJ.tif
MyRegion_region_1_T31TDJ.dbf
MyRegion_region_1_T31TDJ.prj
MyRegion_region_1_T31TDJ.shp
MyRegion_region_1_T31TDJ.shx
MyRegion_region_1_T31TDJ.tif
- ! stats
- statistics
- Optional xml statistics to standardize the data before learning (svm…).
(empty)
- IOTA2_tasks_status.txt
- internal execution status
iota2keeps track of it’s execution using this pickle file (not text) to be allowed to restart from the state where it stopped.
- logs.zip
logs archive
MyRegion.dbf
MyRegion.prj
- MyRegion.shp
- fake region
- When no ecoclimatic region is defined for learning step,
iota2creates this fake file with a single region.
MyRegion.shx
reference_data.dbf
reference_data.prj
- reference_data.shp
- reference data
- ground_truth data where a column “split” has been added
reference_data.shx
Final folder
The final folder contains your prediction maps and csv files with metrics.
The following validation metrics are computed between the reference values in the validation samples and the corresponding predicted values in the final map.
max_errormean_absolute_errormean_squared_errormedian_absolute_errorr2_score
The metrics are computed at the tile and mosaic levels and can be found respectively in the TILE_seed_metrics.csv and mosaic_seed_metrics.csv files.
A summary with the mean and the standard deviation over the different seeds values (when runs argument from section arg_train> 1) is written in TILE_metrics.csv and mosaic_metrics.csv files.
If the merge_run parameter in the multi_run_fusion section is True, you will also find the files Confidence.tif, Regressions_fusion.tif and fusion_metrics.csv.
Confidence.tif and Regressions_fusion.tif are the fusion and confidence maps from the results of the different runs.
The fusion_metrics.csv file gathers the metrics computed on specific samples that are not used to train the models of any run.
Implementation notes
In order to perform the sampling, a fake column named “split” full of zeros is added to ground truth data. The sampling strategy will be applied on this column (see otb sample_selection parameters).
This column is added to the files generated by iota2 and not to the files provided as input by the user.
The target data is standardized before being fed to the model and destandardized after prediction. The scaler is serialized (written to disk) along with the model.
While prediction files have been named with the Regression_TILE_*.tif convention, the folder name classif and some steps named classif_* have been kept for compatibility reasons. Do not be surprised to find things named “classif” even in regression mode.
Available features
Some iota2 features are not available depending on the mode:
currently external features are not available in the
scikit-learnworkflowin the Pytorch workflow, the following features are not supported in regression mode
adaptive learning rate
learning weight
early stop
confidence maps
Known limitations
Regression has some known limitations that remain to be fixed in further contributions:
predictions are done on a whole map. It is not possible to restrict them to given polygons. That’s why validation metrics can only be performed after a full map prediction.
keyword arguments can not be passed to the metric (example HuberLoss
deltaparameter).standardization of targets can not be turned off. It is done in the full target set of samples and can take a lot of memory if the dataset is large.
If you find any other limitations or bugs, please feel free to report them to the iota2 team. The easiest way to do this is to create an issue on framagit describing your comment or bug.
Tutorial
Start with install Iota2 and download the test dataset IOTA2_TUTO_REGRESSION.tar.bz2 (1.5 Go).
You can then use iota2 in regression mode to predict continuous variables. As an example, we provide the reference_ndvi.shp file containing values of NDVI computed by iota2 for pixels in the T31TCJ and T31TDJ tiles.
The sensors images provided are only pieces of tiles to allow a faster execution time.
Include this in your config file (choose the Pytorch or scikit-learn template).
chain:
{
s2_path: "/XXXX/IOTA2_TUTO_REGRESSION/sensor_data"
list_tile: "T31TCJ T31TDJ"
ground_truth: "/XXXX/IOTA2_TUTO_REGRESSION/vector_data/reference_ndvi.shp"
data_field: "ndvi"
}
In the example above, replace the XXXX by the path where the archive has been extracted.
Unlike the Pytorch template the scikit-learn template is configured to perform several executions (runs argument from section arg_train is equal to 3) followed by a merging of the outputs produced.
It is quite possible to use this configuration with a pytorch workflow.
Then run iota2 with the following command:
Iota2.py -config /XXXX/IOTA2_TUTO_REGRESSION/config_tuto_regression.cfg -scheduler_type localCluster
After iota2 has finished, you should find the output directory results_tuto with all the content described in the documentation section Final folder.
If you run the scikit-learn template, you should get the following two outputs, among others:
Here is the fusion map obtained using the scikit-learn model. This image is a single-band raster whose values are the average of the predicted NDVI values between the three runs performed.
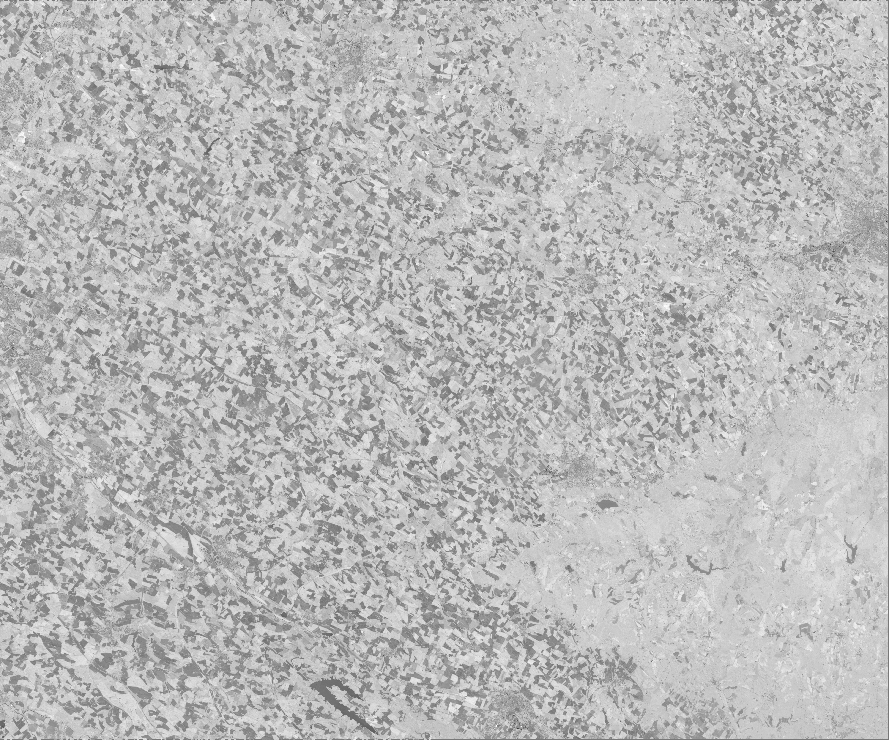
Regressions_fusion.tif
Then we have the table of metrics that are computed on the validation samples for each execution and for the fusion map.
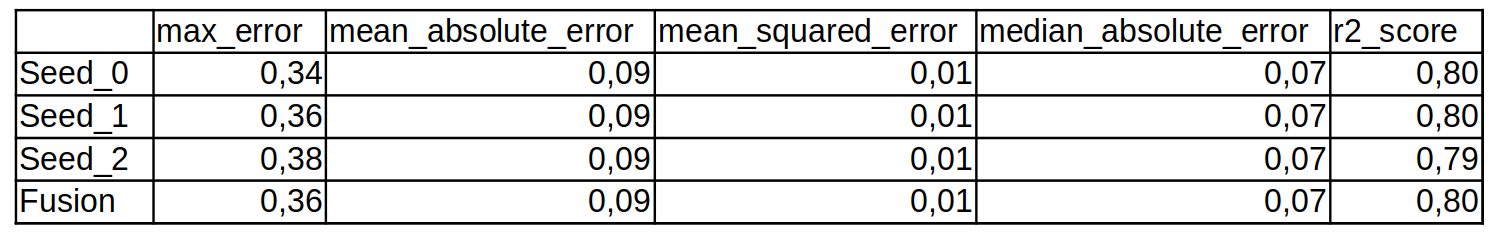
fusion_metrics.csv