iota2’s samples management
This chapter provides explanations about how training and validation samples are managed during the iota2 run. First, we will see how to give iota2 inputs about the dataBase location and some restrictions on it. Next, a focus will be done on the parameters related to the sampling strategy. The different strategies will be illustrated through examples.
Input dataBase location and label’s field
Parameter Key |
Parameter Type |
Default value |
Parameter purpose |
|---|---|---|---|
ground_truth |
string |
Mandatory |
Input dataBase |
data_field |
string |
Mandatory |
Field from the dataBase containing labels |
Note
In the toy data-set data-set, the dataBase file is ‘groundTruth.shp’ and the data_field is ‘CODE’.
Sampling strategy fields
Parameter Key |
Parameter Type |
Default value |
Parameter purpose |
|---|---|---|---|
float |
0.5 |
Range in ]0;1[: ratio between learning polygons / validation polygons |
|
boolean |
True |
Enable the split into learning and validation samples set |
|
integer |
1 |
Number of random splits between learning samples and validation samples, and therefore the number of models and classifications generated |
|
random_seed |
integer |
None |
Random seed: allow iota2 to reproduce random splits between validation and learning sample-set |
integer |
1 |
Threshold to pick learning samples. Every sample which are valid > cloud_threshold can be used to train models |
|
python dictionary |
cf Note 1 |
Sampling strategy in learning polygons |
|
python dictionary |
cf Note 2 |
Generate synthetic samples |
|
string |
None |
Copy samples between samples-set |
Note 1 : sampleSelection default value
sampleSelection:
{
"sampler": "random",
"strategy": "all"
}
which means all pixels inside learning polygons will be used to train models.
Note 2 : sampleAugmentation default value
sample_augmentation:
{
"activate": False
}
which means no sample augmentation will be done.
Different strategies illustrated by examples
Every example is provided with a configuration file allowing users to reproduce outputs.
These configuration files will produce outputs in the '/XXXX/IOTA2_TEST_S2/IOTA2_Outputs/Results'
directory.
split_ground_truth
By default this parameter is set to True. In iota2 outputs, there is a directory named dataAppVal which contains all learning and validation polygons by tile. After a iota2 run, the dataAppVal directory
should contains two files : T31TCJ_seed_0_learn.sqlite and T31TCJ_seed_0_val.sqlite.
Note
files T31TCJ_seed_0_*.sqlite contain polygons for each model, identified by the field region.
See what happens when this parameter is set to False in cfg
As the dataBase input was not split, the two files must contain the same number of features. The entire dataBase is used to learn the model and to evaluate classifications. In pratice this situation should be avoided to reduce the spatial autocorrelation when computing the classification score.
ratio
The ratio parameter allows users to tune the ratio between polygons dedicated to learn models and polygons used to evaluate
classifications when the parameter splitGroundTruth is set to True . By launching iota2 with the ratio parameter cfg
we can observe the content of files T31TCJ_seed_0_*.sqlite in the iota2’s
output directory dataAppVal.
The dataBase input provided groundTruth.shp contains 26 features and 13
different classes. Then by setting the ratio at 0.5, files T31TCJ_seed_0_learn.sqlite
and T31TCJ_seed_0_val.sqlite will contain 13 samples each.
Warning
the ratio is computed considering the number of polygons, not the total area. Also, the ratio is processed by class and by model in order to keep the origin dataBase class repartition.
Note
ratio:0.6 means 60% of eligible polygons will be use to learn models
and 40% to evaluate classifications
runs
This parameter is used to reduce the bias in the estimation of classification accuracy index (OA, Kappa etc…) reported in “RESULTS.txt” (as mentionned in iota2 use case). When it is superior to 1, several runs with random training/validation samples splits are done, and the reported classification accuracies are averaged over the different runs. 95% confidence intervals for estimation of mean OA, Kappa, F-Score, precision and recall are also reported (they are computed with t-test).
cloud_threshold
This parameter allows users to clean-up the dataBase from samples which can not be used to learn models or to evaluate classifications. The pixel validity is used to determine if samples are usable. A valid pixel at time t is a pixel which is not flagged as cloud, cloud shadow or saturated. Thus, usable samples are samples which are valid more than cloud_threshold times.
We can observe the influence of the cloud_threshold parameter by launching iota2 with cfg
First, here is the arborescence the features iota2 output directory
features
└── T31TCJ
├── CloudThreshold_2.dbf
├── CloudThreshold_2.prj
├── CloudThreshold_2.shp
├── CloudThreshold_2.shx
├── nbView.tif
└── tmp
├── MaskCommunSL.dbf
├── MaskCommunSL.prj
├── MaskCommunSL.shp
├── MaskCommunSL.shx
├── MaskCommunSL.tif
└── Sentinel2_T31TCJ_reference.tif
Let’s open nbView.tif and CloudThreshold_2.shp files.
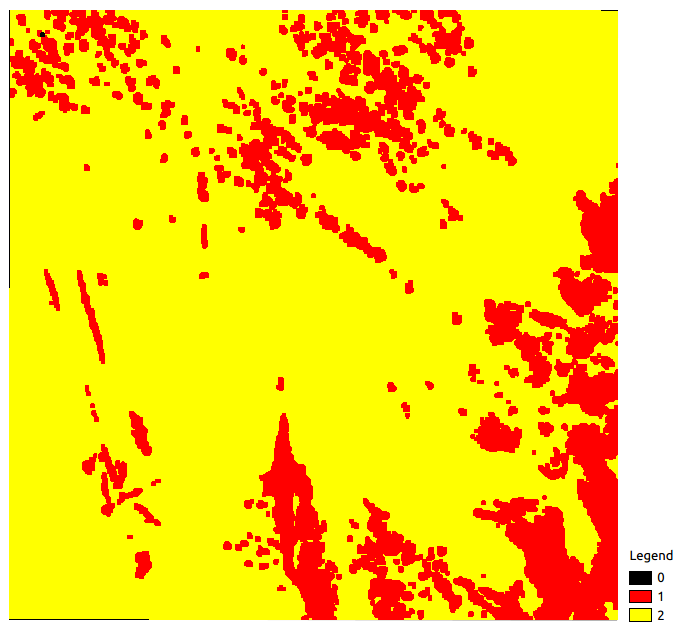
Pixel validity raster |
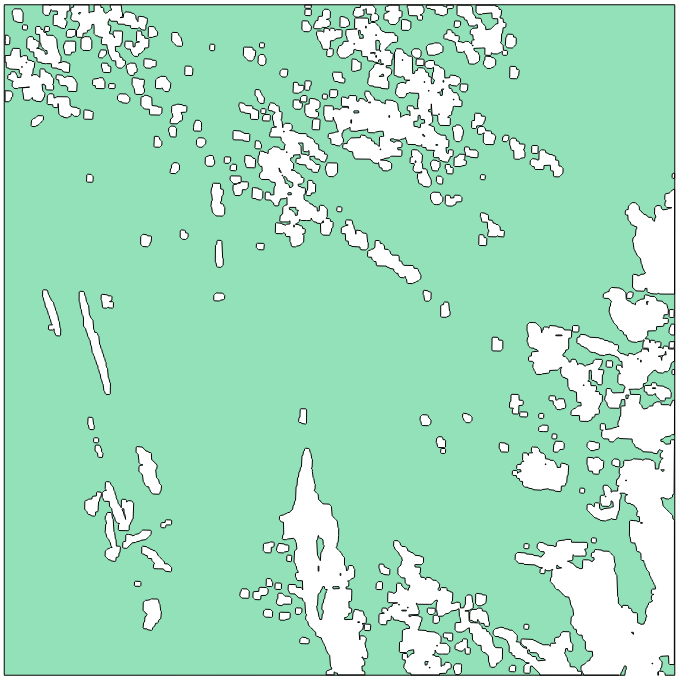
Cloud threshold vector |
As you can notice, every pixel in the validity raster which are superior or equal
to the parameter cloud_threshold value (here 2) belong to a geometry in the
vector file CloudThreshold_2.shp. Next, available polygons are the ones resulting
from the intersection of the CloudThreshold_2.shp vector file and the dataBase input.
sample_selection
Once learning polygons are chosen, it is the time to select pixels by sampling polygons. Many strategies are available through the use of OTB SampleSelection application, this section will detail some of them.
First, we may have a look at the default strategy by using one of previous configuration
file cfg. In order to
visualize the influence of strategies, we can open the file T31TCJ_selection_merge.sqlite
stored in the directory named samplesSelection. Files called *_selection_merge.sqlite
are tile specific and contain every point selected to learn each models and each
seed (random splits).

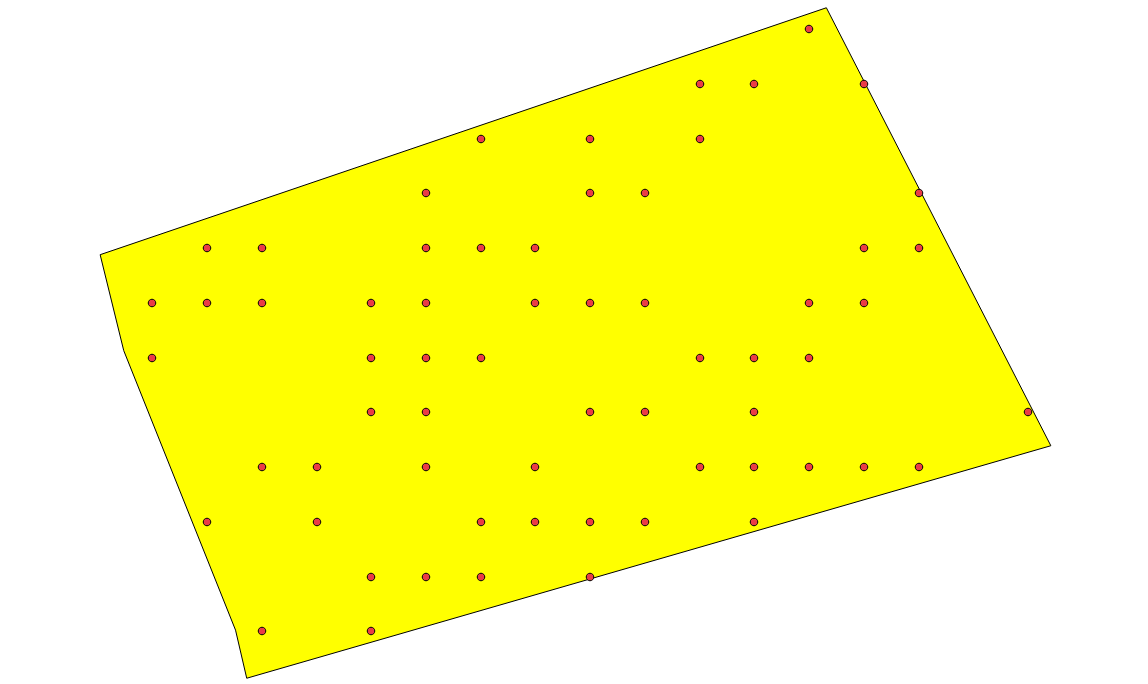
random sampling polygon at 100% rate
Points represent pixel’s centroid selected by the strategy to learn a model. Here, every pixel inside a polygon will be used to learn models. This is the default strategy
sample_selection:
{
"sampler": "random",
"strategy": "all"
}
Sometimes, it may be interesting to change the default strategy to a more suited one (depending on the specific use-case): using High resolution remote sensor, too many polygons, polygons too big, class repartition is unbalanced …
Sampling randomly with a 50% rate
By adding the block below in the configuration file, we use new sampling strategy :
select randomly pixel with a 50% rate. cfg
sample_selection :
{
"sampler":"random",
"strategy":"percent",
"strategy.percent.p":0.5
}
random sampling polygon at 50% rate
Periodic sampling
By changing the sampler argument from random to periodic one pixel
every two is selected for “strategy.percent.p”:0.5. If “strategy.percent.p”:0.1, one pixel every ten pixel would be selected.
sample_selection :
{
"sampler":"periodic",
"strategy":"percent",
"strategy.percent.p":0.5
}
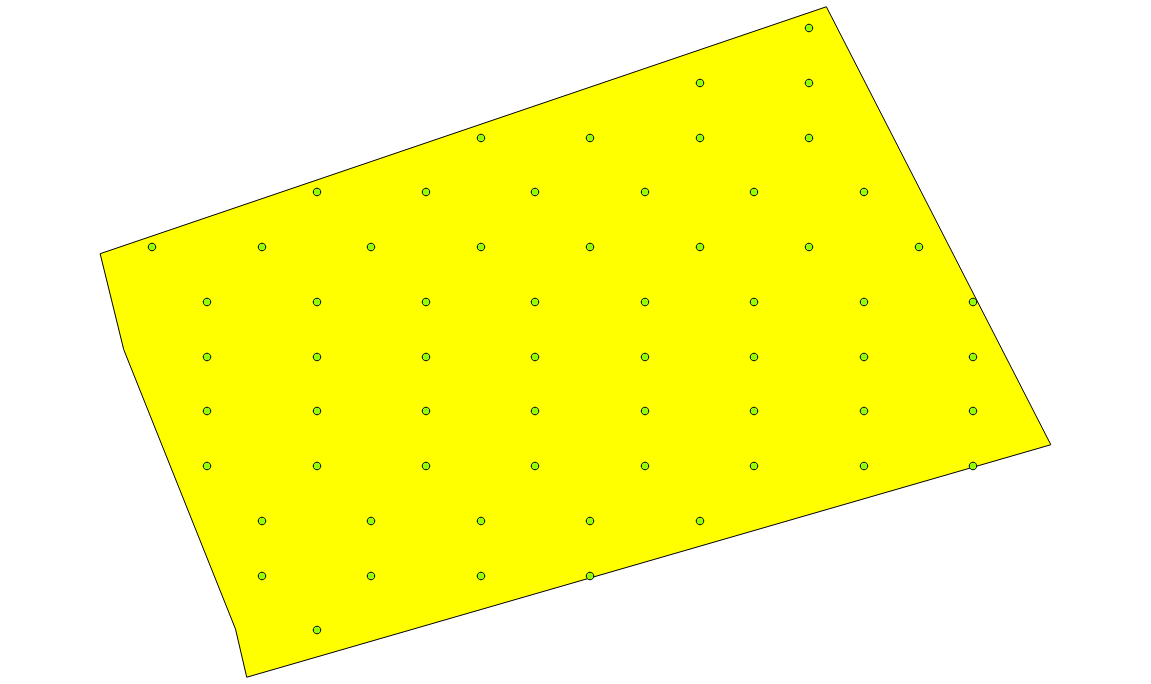
periodic sampling polygon at 50% rate
Different sampling strategy by models
An interesting feature is the ability of iota2 to set a strategy by model.
Obviously, several models must exist and mentionned in the configuration file.
cfg
sample_selection : {"sampler":"random",
"strategy":"all",
"per_model":[{"target_model":2,
"sampler":"random",
"strategy":"percent",
"strategy.percent.p":0.5
}]
}
The aim of this strategy is to sample every polygon with a rate of
100% except polygons belonging to the model 2 which will be sampled with
a 50% rate.
In our case, only two models are invoked, then the strategy presented is equivalent to
sample_selection : {"per_model":[{"target_model":1,
"sampler":"random",
"strategy":"all"
},
{"target_model":2,
"sampler":"random",
"strategy":"percent",
"strategy.percent.p":0.5
}]
}
The argument per_model receives a list of python dictionaries describing a strategy
by target_model. The keys (“sampler”, “strategy”) are the ones provided by
SampleSelection
OTB’s application except target_model which is specific to iota2.
Note
The strategy byclass provided by OTB could also be useful to set
the number of samples selected by class and set ‘manually’ the balance in the
dataBase.
Tracing back the actual number of samples
There are some cases where one wants to know the exact number of samples per class, for example if a classification is run against multiple samples selections with different random seeds. The SamplesSelection OTB App exports one outrates.csv file per model and per seed. When generating the report, those files are aggregated in the learningSamples folder under class_statistics.csv. There is a command line utility allowing to run the aggregation on demand:
# example use
concat_sample_selections.py IOTA2_Outputs/Results_classif/ nomenclature23.txt vector_data/reference_data.shp
wrote csv files to 'learningSamples' folder
# help page
Usage: concat_sample_selections.py [OPTIONS] OUTPUT_PATH NOMENCLATURE REF_DATA_PATH
merge all sample selection files using user label
Options:
-f, --field TEXT original field name in shapefile [default: code]
--help Show this message and exit.
sample_augmentation
Sample augmentation is used to generate sythetic samples from a sample-set. This feature is useful to balance classes in the dataBase. In order to achieve this, iota2 offers an interface to the OTB SampleAugmentation application. To augment samples, users must chose between methods to perform augmentation and set how many samples must be add.
Methods
There are three methods to generate samples : replicate, jitter and smote. The documentation here explains the difference between these approaches.
Number of additional samples
There are 3 different strategies:
- minNumber
To set the minimum number of samples by class required
- balance
Samples are generated for each class until every class reaches the same number of training samples as the largest one.
- byClass
augment only some of the classes
Parameters related to minNumber and byClass strategies are
- samples.strategy.minNumber
minimum number of samples per class.
- samples.strategy.byClass
path to a CSV file containing in first column the class’s label and in the second column the minimum number of samples required.
sample_augmentation’s parameters
Parameter Key |
Parameter Type |
Default value |
Parameter purpose |
|---|---|---|---|
target_models |
list |
Mandatory |
List containing string to target models to augment. target_models : [“all”] to augment all models |
strategy |
string |
Mandatory |
Augmentation strategy [replicate/jitter/smote] |
strategy.jitter.stdfactor |
integer |
10 |
Factor for dividing the standard deviation of each feature |
strategy.smote.neighbors |
string |
Mandatory |
Number of nearest neighbors |
samples.strategy |
string |
Mandatory |
Define how samples will be generated [minNumber/balance/byClass] |
samples.strategy.minNumber |
integer |
Mandatory |
Minimum number of samples |
samples.strategy.byClass |
string |
Mandatory |
path to a CSV file. First column the class’s label, Second column : number of samples required |
activate |
boolean |
False |
flag to activate sample augmentation |
Set augmentation strategy in iota2
sample_augmentation : {"target_models":["1", "2"],
"strategy" : "jitter",
"strategy.jitter.stdfactor" : 10,
"samples.strategy" : "balance",
"activate" : True
}
Here, classes of models “1” and “2” will be augmented to the the most represented class in the corresponding model using the jitter method.:download:cfg<./config/config_samplesAugmentation.cfg>
sample_management
This parameter allows to copy samples from a sample set dedicated to learn a model to another models.
This feature is convenient if a model does not contain enough samples to represent a specific class. Then, the user can provide a CSV file reporting how copy sample by models.
The CSV file must respect the following format :
first column |
second column |
third column |
fourth column |
|---|---|---|---|
source |
destination |
class label |
quantity |
A CSV file containing
1,2,11,2
1,2,31,14
will copy 2 samples (randomly selected) of the class 11 from the model 1 to the model 2. After that, 14 samples of the class 31 will be copied from the model 1 to the model 2.
Note
csv files delimiter must be in : ‘,’, ‘:’, ‘;’ or ‘ ‘ (whitespace)